Figure 1. Down-regulation of MAP3K1 expression by lithium chloride (LiCl).
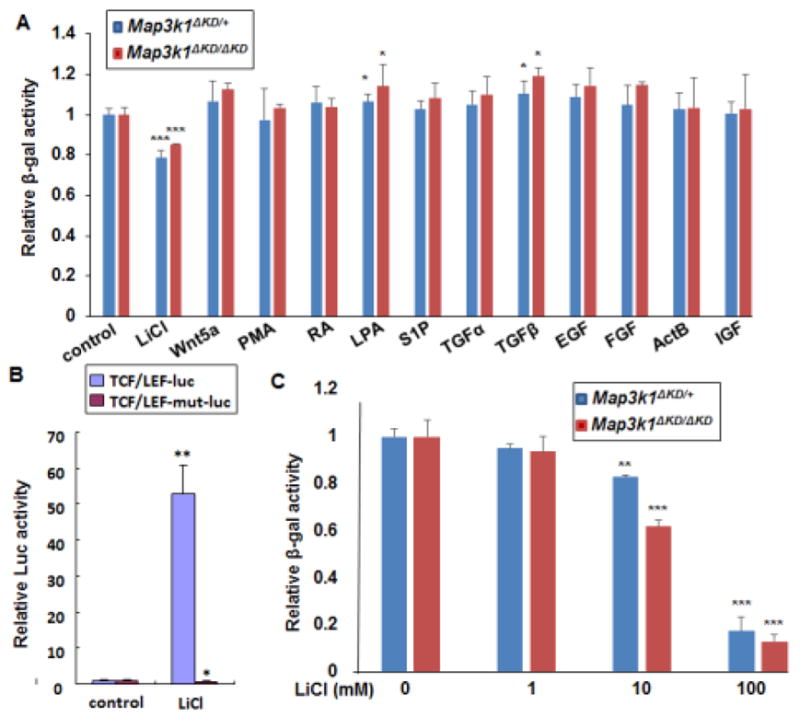
(A and C) Mouse embryonic fibroblasts (MEFs) isolated from mice carrying the knock-in allele, encoding a kinase-dead truncated MAP3K1 protein fused to β-gal (Map3k1ΔKD), as indicated in the figures, were used. The cells were treated for 24 h with (A) different stimuli, including LiCl (10 mM), Wnt5a (100ng/ml), PMA (40nM), RA (10uM), LPA (20uM), S1P (20uM), TGFα (100 ng/mL), TGFβ (10ng/ml), EGF (10 ng/ml), FGF (10 ng/ml), ActB (10 ng/ml) and IGF (100 ng/ml), or (C) various concentrations of LiCl as indicated. The β-gal activities were examined, and relative activities were calculated based on protein concentration. (B) HEK293 cells were co-transfected with CMV-renilla luciferase reporter and Wnt/β-catenin signaling luciferase reporter in its wild type (TCF/LEF-luc) or mutant (TCF/LEF-mut-luc) forms. The cells were treated with LiCl for 24 hours and luciferase activity was examined. The ratio of firefly-luc versus renilla-luc was calculated, and the value in control was set as 1. Statistical significance was determined using Student t-test, * P<0.05, **P<0.01 and ***P<0.001 are considered significantly different from the value in control samples.
