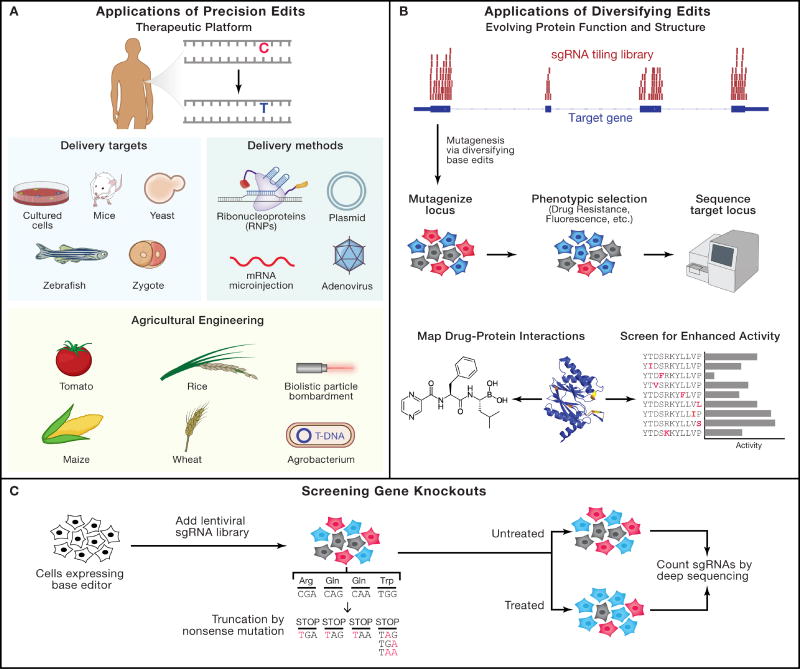
Figure 2. Applications of base editing technologies.
Base editing has been applied in two main areas: precision editing and screening applications.
A) For precision editing, corrective or beneficial conversion of cytidines to thymidines has been demonstrated in many systems. Multiple delivery targets and methods that have been used are shown (see also Table 1). Precision editing has also been used for agricultural engineering using agrobacterium or biolistic DNA particle bombardment to deliver base editors to tomato, maize, wheat, and rice plants.
B) Diversifying base editors have been used to evolve and study protein function and structure. In these experiments, sgRNAs tiling a region are used to generate targeted mutations and the subsequent population of variants is screened for specific phenotypes (i.e. drug resistance/fluorescence), followed by targeted sequencing of the locus. To validate identified mutations, enriched variants are individually installed and drug resistance or the activity of the individual clones are validated.
C) The precise base editing system can also be used for screening applications. Targeting the BE3 system to Arg, Gln, and Trp codons can edit each into stop codons, generating truncated gene products (CRISPR-STOP, iSTOP). This platform provides a potential alternative to Cas9 gene screens without inducing double strand breaks.