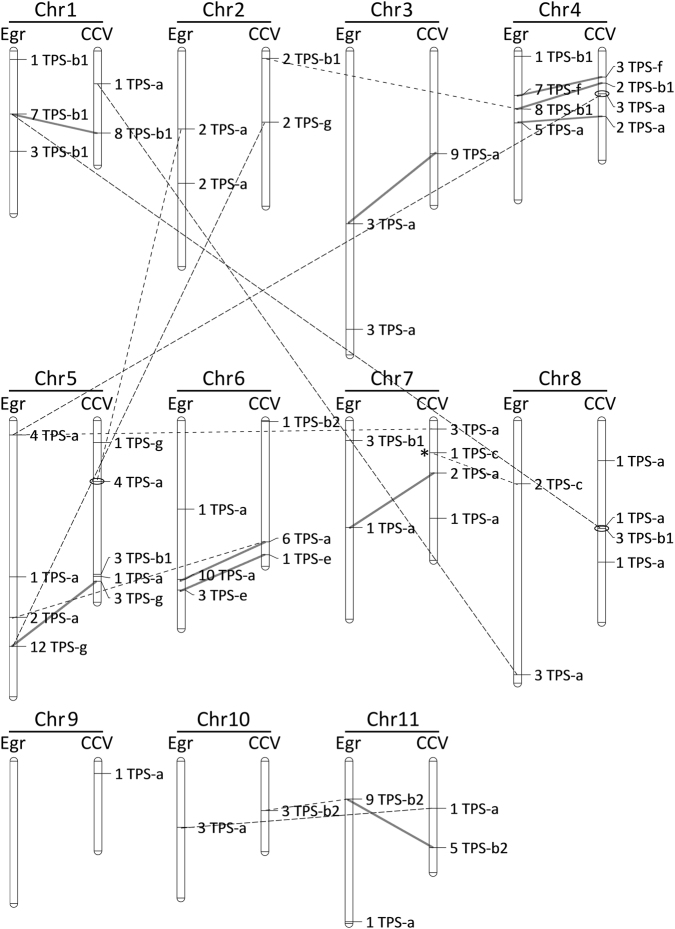
Fig. 4.
Comparison of copy number and genomic location of TPS physical clusters between E. grandis (Egr) and C. citriodora subsp. variegata (CCV). Chromosomes are scaled by physical size. Locus names show the number of TPS genes and the subfamily they belong to. Separate clusters on the same chromosome were defined based on both physical distance and phylogenetic relatedness (see Table S4). Solid lines indicate clusters that are both homologous and syntenic between the two species, while broken lines indicate homologous clusters that are present on different chromosomes in each species. For example, in the TPS-b subfamily, a cluster of eight TPS genes are present on chromosome 4 in E. grandis, in contrast to the syntenic and non-syntenic homologous clusters present in C. citriodora subsp. variegata on chromosome 4 and 2, respectively. Non-syntenic loci between C. citriodora subsp. variegata and E. grandis are circled (only on CCV) to indicate support for this placement based on the CCV54 genome assembly. Similarly, loci are tagged with an asterisk on CCV to indicate disagreement (see Table S5). TPS clusters without lines indicate that their homologue is present in the minor scaffolds of the other species and cannot be examined for synteny. Homology of singleton TPS genes is not shown