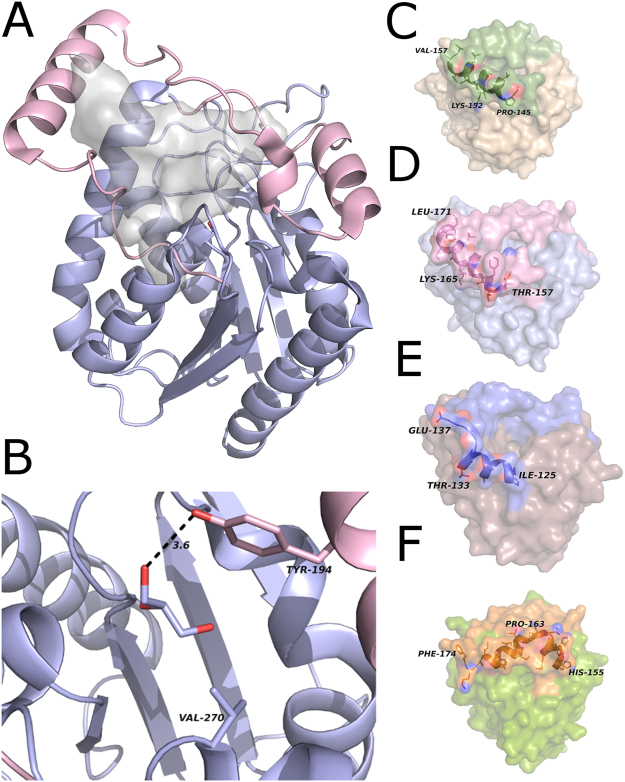
Figure 3.
(A) Overall structure of hMGL (PDB ID 3HJU, chain A). The gray blob shows the cavity inside the protein. The cap is shown in pink and the core in light blue. (B) Close up view of the small cavity in hMGL (PDB ID 3HJU, chain A). The residues at equivalent positions to the glycerol moiety binding residues in mtbMGL are shown as sticks. (C) Surface of mtbMGL with the α/β-hydrolase core in wheat and the cap region in green. The side chains in cap helix 1 shown as sticks and the backbone in cartoon representation. (D) The surface of hMGL with the α/β-hydrolase core in blue and the cap region in pink. The side chains in cap helix 1 are shown as sticks and the backbone as cartoon (PDB ID 3HJU, chain A). (E) Surface representation of bMGL with the α/β-hydrolase core in brown and the cap in blue. The side chains in cap helix 1 are shown as sticks and the backbone as cartoon (PDB ID, 4KEA, chain A).(F) Surface view of the Yju3p structure with the α/β-hydrolase core in green and the cap region in orange. The backbone of cap helix 1 and cap helix 2 is shown as cartoon and the side chains as sticks (PDB ID 4ZWN, chain A).