Abstract
Mapping global parasite diversity is crucial to identify geographical hotspots of emerging disease, and guide public health and conservation efforts. In principle, assuming a bottom-up coupling between the diversity of resources and consumers, the geographical distribution of parasite diversity should match that of host diversity. We test the expected spatial congruence between host and parasite diversity for helminth parasites of vertebrate hosts, across grid cells of a global map. Using high-resolution databases on host species distributions and newly compiled data on the geographical distribution of parasite species discovery, we found positive covariation between host species richness and the number of parasite species discovered, for all vertebrate groups, regardless of the analytical method used, spatial autocorrelation, and spatial resolution. However, all associations were very weak, indicating a poor match between host species richness and parasite species discovery. The research deficit in parasite discovery peaks in areas corresponding to hotspots of host diversity, where disproportionately fewer new parasites are discovered than expected based on local host richness. This spatially biased research effort prevents a full inventory of parasite biodiversity, and impedes predictions of where new diseases may emerge. The host taxon-specific maps we produced, however, can guide future efforts to uncover parasite biodiversity.
Keywords: biodiversity hotspots, global richness, helminths, spatial congruence, species discovery, vertebrate hosts
1. Introduction
As our knowledge of parasite biodiversity expands, increasing efforts are aimed at mapping this diversity in an effort to identify potential geographical hotspots of emerging disease, as well as predict and mitigate the impact of climate change on the distribution of pathogens [1–3]. However, data limitations currently prevent global biodiversity maps from being established for most parasite taxa. From first principles, we might expect parasite biodiversity maps to match those of their hosts. In consumer–resource interactions, local consumer species richness is often driven by local resource richness in a bottom-up fashion [4,5]. On regional scales, several studies have indeed reported a positive relationship between host richness and parasite richness across well-sampled local habitat patches [6–8]. A meta-analysis has confirmed that this relationship is very strong and universally observed across various host and parasite taxa [9]. Assuming this trend can be extrapolated to larger spatial scales, we may thus predict that for any given group of parasites and for any given level of host specificity, the global distribution of parasite biodiversity should be a rough mirror image of that of their hosts.
Empirical validation of this simple prediction is presently not possible, but should become feasible in the near future given the increasing rate at which new parasite species are discovered and described [1,10,11]. Parasite species discovery provides a window into parasite diversity, but the former is not necessarily a good proxy for the latter. It could be, presuming the search effort for new parasite species was spatially proportional to host diversity. Hotspots of host diversity are a priori the best areas to prospect for new parasite species, and should therefore be key targets of parasite discovery effort. However, research in ecology and systematics is known to be spatially biased, with researchers exerting disproportionate efforts toward the study of the regional biota near their home institution [12–14]. The concentration of universities and museums with research-active staff is lower in geographical areas of high biodiversity, i.e. in the tropics, than in areas at higher latitudes, especially Europe and North America. As a consequence, the discovery and description of new parasite species in various regions may not occur at a rate directly proportional to host richness in those regions.
We test the hypothesis that the global distribution of parasite discovery effort shows some, but only weak and limited, geographical congruence with host species richness. For this, we focus on the four main groups of parasitic worms of vertebrates: Cestoda, Trematoda, Nematoda and Acanthocephala. For each vertebrate class, we estimate the level of global congruence between the distributions of host richness and parasite records and identify the most understudied geographical regions in terms of parasite discovery, and therefore the regions where future search efforts for parasites are most likely to be fruitful and lead to gain in biodiversity knowledge. We also explore potential differences in sampling effort among vertebrate classes to see whether these can account for differences in patterns of parasite species discovery. Our results thus provide a guide for the optimization of future species discovery and taxonomic efforts.
2. Material and methods
(a). Data collection
We compiled data on the spatial distribution of parasite species discovery from species description records by conducting a detailed search on the ISI Web of Science™ for the period of 1970–2017, as numbers of parasite species discovered and described annually have been much higher in the past 50 years than ever before [1]. The search was restricted to acanthocephalan, cestode, trematode and nematode parasites of vertebrates (see electronic supplementary material for details). Species redescriptions were also considered whenever the original description was made prior to 1970 and if based on new material. We examined all retrieved publications individually and recorded from all genuine species descriptions: (i) parasite species name, (ii) higher taxon, (iii) description type (i.e. new or redescription), (iv) host species, (v) host higher taxon, (vi) habitat, i.e. terrestrial, freshwater or marine, (vii) locality where the parasite was discovered, (viii) its latitude and longitude, and (ix) the full reference. Whenever geographical coordinates were not given in the original article, locality coordinates were obtained from Google Earth v. 7.3.0 [15]. The final dataset included 4889 articles, from which descriptions of 4943 parasite species were collected (table 1).
Table 1.
Global numbers of host and parasite species analysed, number of parasite species that were redescriptions, and number of cells occupied and percentage of occupancy at both spatial resolutions.
| resolution = 1° |
resolution = 2° |
||||
|---|---|---|---|---|---|
| no. species (redescriptions) | no. cells | % occupancy | no. cells | % occupancy | |
| amphibians | 6490 | 14 082 | 21.73 | 3947 | 24.36 |
| all parasites | 241 (25) | 164 | 0.25 | 149 | 0.92 |
| Acanthocephala | 15 (4) | ||||
| Cestoda | 16 (1) | ||||
| Trematoda | 57 (10) | ||||
| Nematoda | 153 (10) | ||||
| reptiles | 4567 | 38 199 | 58.95 | 9682 | 59.77 |
| all parasites | 490 (66) | 307 | 0.47 | 259 | 1.60 |
| Acanthocephala | 11 (0) | ||||
| Cestoda | 49 (5) | ||||
| Trematoda | 110 (23) | ||||
| Nematoda | 320 (38) | ||||
| birds | 8503 | 57 160 | 88.21 | 14 367 | 88.69 |
| all parasites | 609 (112) | 349 | 0.54 | 284 | 1.75 |
| Acanthocephala | 46 (9) | ||||
| Cestoda | 164 (33) | ||||
| Trematoda | 229 (35) | ||||
| Nematoda | 170 (35) | ||||
| terrestrial mammals | 5303 | 24 411 | 37.67 | 6483 | 40.02 |
| all parasites | 1129 (171) | 647 | 1.00 | 527 | 3.25 |
| Acanthocephala | 18 (3) | ||||
| Cestoda | 231 (33) | ||||
| Trematoda | 111 (12) | ||||
| Nematoda | 769 (123) | ||||
| freshwater fish | 6410 | 15 024 | 23.19 | 4249 | 26.23 |
| all parasites | 727 (132) | 383 | 0.59 | 304 | 1.88 |
| Acanthocephala | 71 (7) | ||||
| Cestoda | 142 (50) | ||||
| Trematoda | 235 (27) | ||||
| Nematoda | 279 (48) | ||||
| marine fish | 3657 | 38 381 | 59.23 | 9936 | 61.33 |
| all parasites | 1747 (242) | 581 | 0.90 | 452 | 2.79 |
| Acanthocephala | 83 (11) | ||||
| Cestoda | 551 (75) | ||||
| Trematoda | 793 (113) | ||||
| Nematoda | 320 (43) | ||||
For data on host species richness, we downloaded from the IUCN online database (http://www.iucnredlist.org/technical-documents/spatial-data) [16] data on species' geographical distributions of amphibians, reptiles, terrestrial mammals, freshwater and marine fishes (including both Osteichthyes and Chondrichthyes). Note that IUCN data on reptiles, marine fish and freshwater fish are considered ‘not comprehensive’. For birds, data were obtained from BirdLife International [17] with permission for their non-commercial use. The original providers of the vertebrate host data remain the owners of the data.
(b). Spatial analysis
To generate global maps of both parasite discoveries and host species richness, species' geographical distribution data were transformed into two presence–absence matrices, one with a global grid of 1° of resolution and the other with 2° resolution, using the function lets.presab of the R package letsR [18]. To explore similarity (or dissimilarity) in patterns of spatial distribution between parasite species discoveries and host species richness, we computed correlation coefficients among grid cells, separately for all the six vertebrate groups, at each of the two resolutions. Given the sample sizes for each of the four parasite groups, calculations were performed only for the pooled parasite data. Prior to statistical analysis, joint absences (double zeros, i.e. grid cells where hosts do not occur and no parasite has been found) were excluded from the dataset, since they artificially contribute to similarity between variables [19,20]. We first computed Spearman's correlations (R function cor.test), ignoring spatial autocorrelation. However, species distributional data often display spatial autocorrelation, i.e. locations close to each other are more likely to have comparable values than expected by chance [21]. Both host and parasite species distribution data are likely to be spatially autocorrelated, which to some degree can result from sampling biases especially in the case of parasite species discovery. Statistically, this lack of independence means that each sampling location does not represent a full degree of freedom, and adjusted degrees of freedom should be used to account for the intensity of spatial autocorrelation in each variable. Given the scale of our study, to control for spatial non-independence we used a modified t-test [22] to calculate the statistical significance of the correlation coefficient (a corrected Pearson's correlation) based on geographically effective degrees of freedom [23] as implemented in the SpatialPack package (function modified.ttest) [24]. Since the reliability of this correction is directly related to the estimated degree of spatial autocorrelation, which in turn varies according to the number of distance classes [25,26], the correlation was calculated for five, 13 (default) and 20 classes. To examine patterns of autocorrelation of each variable, the estimated Moran's indices [27] of each variable (also an output from the function modified.ttest) were plotted as a correlogram.
To more explicitly consider spatial information when determining the degree of association between the distributions of parasite discoveries and host species richness, we calculated the Tjøstheim's coefficient [28] with the function cor.spatial (SpatialPack package) [24]. The codispersion coefficient (also known as Matheron's coefficient; [29]) which quantifies the coefficient of association between two spatial variables that are separated by a distance h (lags) was also estimated using the function codisp of the SpatialPack package for up to 13 distance classes. The above measurements tackle different aspects of spatial correlation, with codispersion and the corrected Pearson's correlation coefficient being more similar (see [30–32] for further discussion).
To visually represent the mismatch between the global distribution of parasite discoveries and that of host species richness while accounting for differences in study effort, we first obtained relative values by dividing the raster containing numbers of species per cell by the total number of species of either parasites found or known hosts, for each of the two resolutions. Then, we subtracted the relative value for hosts from that for parasites of the same cell, across all cells, and produced global maps with the resulting values. A predominance of values very close to zero, either negative or positive, would indicate strong proportionality between local host species richness and how many parasite species have been found. The higher the resulting value in a cell (the more positive it is), the greater the relative discovery of parasites relative to the local host species richness. Conversely, cells with low resulting values (i.e. strongly negative values) represent areas where disproportionately few parasites have been discovered relative to local host richness.
Also, we examined whether differences in sampling effort among host groups shape patterns of parasite species discovery. We calculated the percentage of total known host species richness (from IUCN and BirdLife International data) represented by the host species in our database, i.e. hosts from which new parasites have been discovered between 1970 and 2017. We also calculated the percentage of host species in the database from which more than one parasite was described (i.e. host sharing). Typically, from a sample of individual hosts taken from one population (one grid cell), only one new parasite species is described. However, sometimes two or more parasites are described from the same host sample. To test whether the relationship between the number of parasite species described per grid cell and the number of host species from which parasites were found varies among vertebrate host groups, we used spatial generalized linear mixed models (GLMM). To account for spatial autocorrelation, we fitted the structure of the variance–covariance matrix to the data as described in [21] (see electronic supplementary material for details). Coefficients were computed as odds ratio, such that they represent the slope of the relationship, with a value of 1 indicating that for each host species sampled in a grid cell, one parasite species was described. Analyses were performed separately for the six vertebrate host groups (amphibians, reptiles, birds, terrestrial mammals, freshwater fish, marine fish) by pooling all parasite species, and separately for the four parasite groups (Acanthocephala, Cestoda, Nematoda and Trematoda) pooling all host taxa.
Finally, to visualize how parasite species discovered accumulate as a function of the number of sampled grid cells for each vertebrate and parasite taxon, we calculated the cumulative sum of parasite species (excluding cells where zero parasites were found; R function cumsum) with each additional cell sampled of a global grid of 2° of resolution, as described above. We computed 999 random permutations of the order of cells to obtain a 95% confidence interval. All analyses were performed in the R statistical computing environment [33].
3. Results
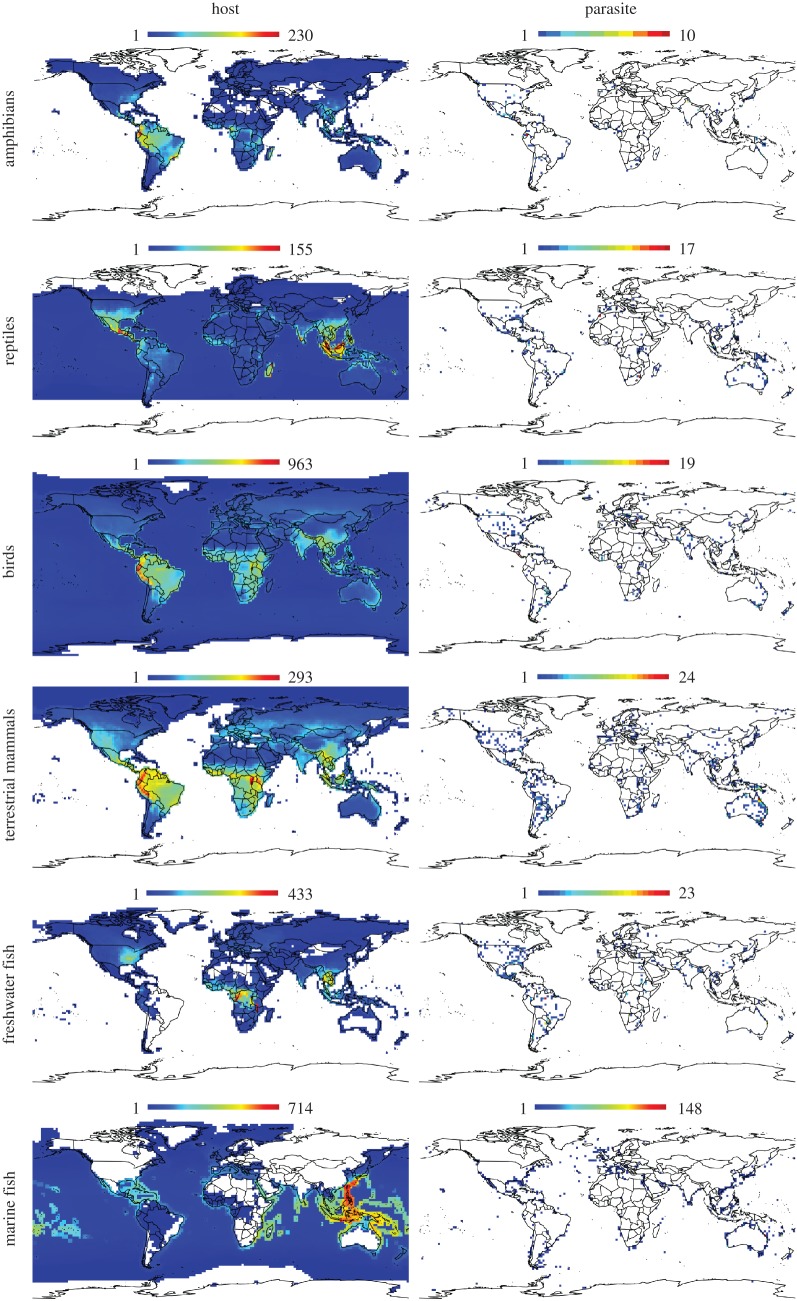
In the five decades covered by our search, nematodes were the most frequently discovered parasite taxon of vertebrates, except for birds and marine fish where trematodes were the most common parasite group (table 1). In all cases, the number of parasite species discovered was always lower, often by an order of magnitude, than the species richness of their respective hosts, whether globally or on a cell-by-cell basis (figure 1 and table 1). Also in all cases, the number of grid cells in which parasites have been found is always a tiny fraction of that known to be occupied by hosts (table 1).
Figure 1.
Global richness maps for amphibians, reptiles, birds, terrestrial mammals, freshwater and marine fish, as well as for the discoveries of their respective parasites. All maps have 2° of resolution. Colour gradients scale linearly with species number per cell. Note that birds and reptiles include ocean-dwelling species. (Online version in colour.)
Overall, for all six groups of vertebrates and at both resolutions, spatial associations between host species richness and parasite species discovery were always positive and statistically significant at a global scale, but very weak (table 2). The strength of each correlation increases only slightly if double zeros are included (data not shown). The degree of association was always a little higher when calculated at a coarser resolution (2° × 2° grid cells; table 2). The values of Spearman's correlations (which ignores spatial autocorrelation), corrected Pearson's correlations and codispersion coefficients were in all cases comparable, while the Tjøstheim's coefficient was the least congruent (table 2). Discrepancies were especially noticeable for associations where one or both variables had a weak spatial autocorrelation as estimated with Moran's index (electronic supplementary material, figures S1 and S2). We did not detect any difference in significance of the corrected Pearson's correlation coefficient across the three distance classes. The codispersion coefficient was positive for all the lags indicating a match between host and parasite occurrences, while still varying across the different distance lags (electronic supplementary material, figure S3). The association between richness of terrestrial mammals and the number of their parasites discovered per cell showed the highest global congruence when spatial autocorrelation was not considered and also based on Tjøstheim's coefficient. In contrast, based on the Pearson's correlation corrected for spatial autocorrelation and codispersion coefficients, the association between reptiles and their parasites had the highest global congruence. The association between freshwater fishes and their parasites showed the lowest global congruence across all coefficients (table 2).
Table 2.
Degree of spatial association between host species richness and parasite species discovery for different vertebrate host groups, based on Spearman's correlation, the modified t-test (p-values shown for five, 13 and 20 distance classes), Tjøstheim's coefficient, and the codispersion coefficient for the maximum value with the respective distance lag (h).
| Spearman's |
modified t-test |
Tjøstheim's |
codispersion |
||||||
|---|---|---|---|---|---|---|---|---|---|
| resolution | no. cells | rho | p | cor | p | cor | variance | max | distance h |
| amphibians | |||||||||
| 1° | 14 082 | 0.1008 | *** | 0.1124 | ***/***/*** | 0.1235 | 5.14 × 10−5 | 0.1235 | 13 |
| 2° | 3947 | 0.1858 | *** | 0.2099 | ***/***/*** | 0.0850 | 1.85 × 10−4 | 0.2337 | 9 |
| reptiles | |||||||||
| 1° | 38 199 | 0.1300 | *** | 0.1547 | ***/***/*** | 0.0285 | 2.21 × 10−5 | 0.1695 | 13 |
| 2° | 9682 | 0.2305 | *** | 0.2480 | ***/***/*** | 0.0197 | 8.69 × 10−5 | 0.2699 | 13 |
| birds | |||||||||
| 1° | 57 160 | 0.1056 | *** | 0.0935 | ***/***/*** | 0.0238 | 1.27 × 10−5 | 0.1013 | 13 |
| 2° | 14 367 | 0.1865 | *** | 0.1603 | ***/***/*** | 0.0338 | 5.05 × 10−5 | 0.1741 | 13 |
| terrestrial mammals | |||||||||
| 1° | 24 412 | 0.1592 | *** | 0.1176 | ***/***/*** | 0.1937 | 2.53 × 10−5 | 0.1278 | 12 |
| 2° | 6484 | 0.2792 | *** | 0.1882 | ***/***/*** | 0.1968 | 9.66 × 10−5 | 0.2039 | 12 |
| freshwater fish | |||||||||
| 1° | 15 089 | 0.0272 | *** | 0.0545 | ***/*/* | 0.0396 | 4.44 × 10−5 | 0.0924 | 3 |
| 2° | 4296 | 0.0719 | *** | 0.0895 | ***/*/* | 0.0470 | 1.56 × 10−4 | 0.1573 | 3 |
| marine fish | |||||||||
| 1° | 38 391 | 0.1401 | *** | 0.0803 | ***/***/*** | 0.0242 | 2.08 × 10−5 | 0.0952 | 7 |
| 2° | 9946 | 0.2280 | *** | 0.1393 | ***/***/*** | 0.0180 | 7.97 × 10−5 | 0.1599 | 7 |
***p < 0.001, **p < 0.01, *p < 0.05.
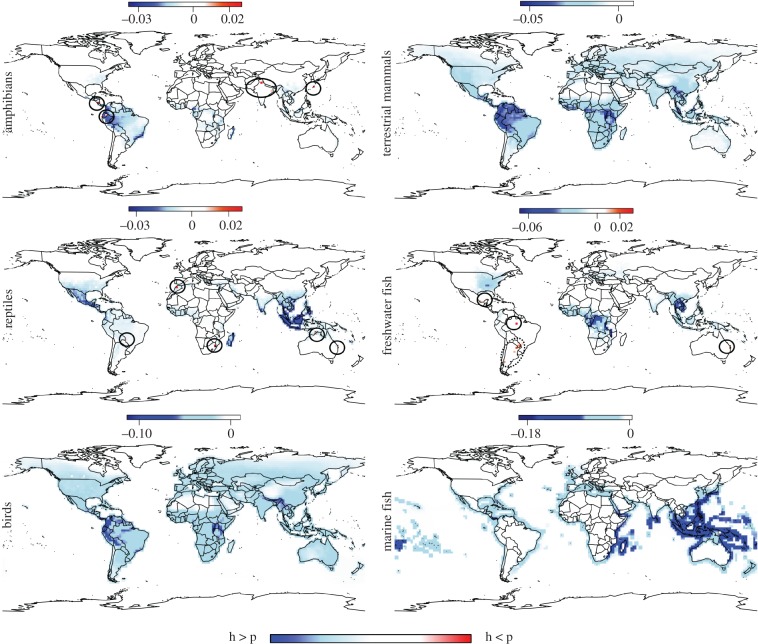
We observed very poor congruence between the relative parasite discovery effort and relative host species richness, for all host taxa, with within-cell difference values being overwhelmingly negative (figure 2). In other words, in the majority of geographical areas, disproportionately fewer parasite species have been discovered than expected based on local host richness. For amphibians, reptiles and freshwater fish, some grid cells presented relative numbers of parasite species discovered higher than the local relative host species richness, while for the other vertebrate groups this was never the case. The few cases of relatively higher parasite species discovery (positive values) did not correspond to cells with high host species richness, with the exception of an area in Ecuador for amphibians (figures 1 and 2). In fact, parasite species descriptions have only been made for less than 3% of the known species richness of amphibians, and up to about 28% of the species richness of marine fish recorded in the IUCN database (table 3). However, the latter estimate is probably an overestimate given that the IUCN data for marine fish is considered ‘not comprehensive’.
Figure 2.
Global distribution maps of relative parasite discovery effort compared to relative host species richness at 2° of resolution. Colour gradients represent differences in proportionality of local host species richness and how many parasite species have been found: blue cells represent areas where disproportionately few parasites have been discovered relative to local host richness (h > p); white cells represent values close to zero indicative of strong proportionality between local host species richness and how many parasite species have been found; red grid cells represent values above zero indicating a greater discovery of parasites relative to the local host species richness (h < p). The few red cells are also highlighted with a black circle. Dashed ellipse represents an area where more parasites have been found but no host distribution data are currently available from the IUCN database [16]. (Online version in colour.)
Table 3.
Sampling effort, measured as the percentage of the known richness of the group from which parasites have been discovered (% Rich.), and degree of host sharing for each host and parasite group. Estimates of the relationship between the number of parasite species described per grid cell and the number of host species from which parasites were found are from generalized linear mixed models, and given in odds ratios for 2 degrees of cell resolution.
| no. cells | parasite species | host species (% Rich.) | host sharing (%) | estimates | p | |
|---|---|---|---|---|---|---|
| amphibians | 149 | 241 | 160 (2.47) | 33.6 | 1.3240 | *** |
| reptiles | 259 | 490 | 324 (7.09) | 33.9 | 1.5300 | *** |
| birds | 271 | 567 | 369 (4.34) | 34.9 | 1.2825 | *** |
| terrestrial mammals | 522 | 1117 | 601 (11.33) | 46.2 | 1.3150 | *** |
| freshwater fish | 304 | 727 | 477 (7.44) | 34.4 | 1.2202 | *** |
| marine fish | 452 | 1746 | 1013 (27.70) | 42.0 | 1.0512 | *** |
| Acanthocephala | 165 | 244 | 224 | 8.2 | 1.2319 | *** |
| Cestoda | 615 | 1154 | 747 | 35.2 | 1.2569 | *** |
| Nematoda | 800 | 2011 | 1382 | 31.2 | 1.1342 | *** |
| Trematoda | 498 | 1485 | 1001 | 32.6 | 1.0553 | *** |
***p < 0.001.
Host sharing, i.e. the percentage of host species in the database from which more than one parasite was described, was very similar across all vertebrate host groups (table 3). Values for host sharing range between 33% and 46% among host groups, though this is across the dataset, not within cells. Not surprisingly, for each of the six vertebrate groups and for each parasite group, the number of host species sampled per grid cell significantly predicted the number of parasite species discovered per grid cell (in all cases, p < 0.001; table 3). However, in some cases variograms and correlograms (Moran's I) indicate some degree of spatial autocorrelation in model residuals (data not shown). Coefficients (odds ratio) obtained from the GLMMs are all slightly greater than 1, with the maximum value observed for reptiles (table 3), meaning that across grid cells only one parasite is usually described per sampled host species, with only rare exceptions. The similarity among coefficients (table 3) indicates that the discovery rate of new parasite species per host sampled is similar across the six vertebrate host groups. The cumulative numbers of parasite species increased linearly as a function of the number of grid cells sampled (electronic supplementary material, figure S4). Differences among taxa are due to differences in the total number of parasites discovered and in the maximum number found per cell. The overlapping confidence intervals indicate that parasite discovery versus sampling effort follows the same pattern across vertebrate groups, except for marine fishes, where new parasite species accumulate a little faster with each additional cell sampled than for other vertebrates. The cumulative rise in numbers of parasite species as a function of the number of grid cells sampled was also similar among parasite taxa when these were pooled regardless what kind of host they came from (electronic supplementary material, figure S4).
4. Discussion
A robust biogeography of parasites is essential to underpin global public health and conservation initiatives [34,35]. However, our discovery and compilation of parasite species is lagging far behind our knowledge of vertebrate diversity and distribution [1]. Although there exist methods to estimate the ‘missing’ parasite species richness in undersampled regions [3], these are poor substitutes for actual lists of identified species. Here, we provide the first global maps of the known diversity of helminth parasites, based on where they have been discovered. Our literature search was time-limited (1970–2017) and most likely missed several parasite species descriptions, but nevertheless provided a robust dataset from which to draw those maps. Although our data indicate that parasites have only been reported from a small fraction of areas where hosts occur and for a small percentage of known host species, we demonstrate convincingly that regardless of the statistical method or the spatial resolution used, the number of parasite species discovered covaries positively and significantly with local host species richness, for all vertebrate host taxa.
The strength of these associations, however, is very weak. On smaller spatial scales, the covariation between local parasite species richness and host species richness among well-sampled habitat patches is universally strong across all available studies on various host and parasite taxa (average effect size in meta-analysis = 0.55) [9]. This strong bottom-up control of parasite richness by available host richness is scale-independent: although the spatial scale (maximum distance between habitat patches) varied widely across studies, from 10 to 104 km, the strength of the host richness–parasite richness relationship was unaffected by study scale [9]. In contrast, on a global scale, here we only found correlation values less than 0.25, often less than 0.10, indicative of very weak relationships. There is no biological reason to expect such weak associations on this spatial scale. The global distribution of vertebrate species richness is relatively well known and the host data we used are likely a true representation of real vertebrate diversity; therefore, the problem must be with the parasite data. Because the number of parasite species discovered per grid cell across the five decades covered by our dataset is a direct reflection of research effort in parasite biodiversity and taxonomy, we can infer that parasite diversity has been particularly understudied in areas of highest host species richness.
Indeed, the research deficit in parasite discovery is apparent in most geographical areas, but peaks in areas corresponding to hotspots of host species richness (compare figures 1 and 2). Even on the Great Barrier Reef, where research on the diversity of parasites (especially trematodes) of marine fish has been intense in recent decades [36], the overall number of parasite species discovered is still disproportionately low relative to local host richness. There are some rare exceptions: small areas where a disproportionately high number of parasite species have been found relative to the local species richness of amphibians, reptiles or freshwater fish. These are probably the result of exhaustive parasite surveys with unusually broad coverage of local host species. Only one of these cases (parasites of amphibians in Ecuador) occurs in a hotspot of host diversity. Generally, areas of high host species richness have yielded disproportionately few new parasite species. Not surprisingly, they remain the most promising regions to prospect for new parasite species. Most likely, the limited search for parasite species in tropical hotspots of host diversity is due to the fact that researchers in ecology and systematics, most of whom are based in temperate regions, direct their efforts mainly toward the study of the regional biota near their home base [12–14]. This geographical bias is a major impediment toward the completion of the global parasite species inventory [37] and its use in conservation biology and disease management [34,35]. In addition, there is a taxonomic bias in parasite discovery effort, with a relatively higher proportion of known host species in some groups (mammals and fish) having been sampled for helminth parasites than other vertebrate groups.
The weak but significantly positive association we found between local host species richness and the number of parasite species reported is robust across different analytical approaches. The discrepancies we observed in some cases between estimates of the Tjøstheim's coefficient and the other coefficients were probably a consequence of the weak spatial trend in the parasite data (table 2; electronic supplementary material, figures S1 and S2). Indeed, Tjøstheim's coefficient often fails to detect existing associations in the absence of sufficient spatial autocorrelation in the variables [30]. The degree of congruence observed between host species richness and parasite discovery was also influenced by the spatial resolution of the analysis, being stronger at 2° than at 1° resolution. Spatial congruence is often resolution-dependent, becoming weaker at finer resolution ([38], but see [39]). However, given the nature of host–parasite associations in which ecological interactions are scale-dependent, mismatches may occur when the chosen scale mischaracterizes the actual spatial patterns of congruent diversity. Nevertheless, our results are similar whether we performed the analyses at 2° or at 1° resolution.
The ecological characteristics of parasites may affect parasite discovery rates. For example, all else being equal, a generalist parasite that infects many host species is more likely to be discovered before a specialist parasite [40]. Similarly, host sharing, i.e. host species harbouring more than one parasite species, may increase the number of parasite species described per grid cell. However, the reality of parasite species descriptions highlights an important sampling bias, such that the host sharing we measured is actually the proportion of host species from which more than one parasite species were described. Even if a host sample yields more than one parasite species during dissection, most species are usually neither described nor reported. The high host sharing values we find in our dataset indicate that often several parasite species are discovered from the same host species, but across its entire geographical range, not within a grid cell. Our results show that the number of parasite species discovered scales positively with either the number of host species sampled or the number of grid cells sampled, roughly equally across vertebrate host taxa or across helminth taxa. Marine fishes are the exception, as the number of parasite species rises more steeply with each new grid cell sampled than for other vertebrate groups. This may simply be due to the greater diversity of fish (globally and per grid cell), and the fact that more of them have been sampled for parasites, in both absolute and relative terms (table 3). Overall, these findings suggest that at a global scale, the majority of descriptions are made as one parasite species per host species per sampling locality. Importantly, this inefficient sampling approach for parasite biodiscovery applies roughly equally across all host or parasite taxa.
In conclusion, we provide the first global maps of the spatial distribution of known parasite diversity, based on the locations of their discovery. Our results indicate that parasite discovery effort shows only a poor geographical match with host species richness; in particular, new parasite species are found disproportionately less often in hotspots of host diversity than expected based on their host species richness. These findings suggest that parasite discovery efforts lag well behind our knowledge of host diversity, and cast doubts on previous attempts to extrapolate global parasite biodiversity [37,41]. However, the maps generated here (especially figure 2) provide clear guides for future research effort by identifying areas suffering from a deficit in parasite species prospecting, and thus the most promising areas in which to seek new parasites.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank BirdLife International and the IUCN for permission to use data on vertebrate species distributions. We also thank P. Tarroso for assistance with the cumulative analysis.
Ethics
This study did not require ethical approval because no animals were handled. Analysed data were obtained from published studies and from IUCN and BirdLife International with permission for their non-commercial use. The original providers of the vertebrate host data remain the owners of the data.
Data accessibility
Species distribution data are available in polygon format on the IUCN Red List of Threatened Species database (http://www.iucnredlist.org/technical-documents/spatial-data), and from BirdLife International (http://datazone.birdlife.org/species/requestdis). The full dataset on the geographical distribution of parasite species descriptions and the R script used in the analysis are uploaded as electronic supplementary material.
Authors' contributions
R.P. and F.J. designed the study; F.J. compiled the data and conducted the analyses; both authors equally shared the writing of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by a University of Otago Research Grant to R.P.
References
- 1.Poulin R. 2014. Parasite biodiversity revisited: frontiers and constraints. Int. J. Parasitol. 44, 581–589. ( 10.1016/j.ijpara.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 2.Estrada-Peña A, Ostfeld RS, Peterson AT, Poulin R, de la Fuente J. 2014. Effects of environmental change on zoonotic disease risk: an ecological primer. Trends Parasitol. 30, 205–214. ( 10.1016/j.pt.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 3.Stephens PR, et al. 2016. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 19, 1159–1171. ( 10.1111/ele.12644) [DOI] [PubMed] [Google Scholar]
- 4.Scherber C, et al. 2010. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468, 553–556. ( 10.1038/nature09492) [DOI] [PubMed] [Google Scholar]
- 5.Castagneyrol B, Jactel H. 2012. Unraveling plant–animal diversity relationships: a meta-regression analysis. Ecology 93, 2115–2124. ( 10.1890/11-1300.1) [DOI] [PubMed] [Google Scholar]
- 6.Krasnov BR, Shenbrot GI, Khokhlova IS, Degen AA. 2004. Relationship between host diversity and parasite diversity: flea assemblages on small mammals. J. Biogeogr. 31, 1857–1866. ( 10.1111/j.1365-2699.2004.01132.x) [DOI] [Google Scholar]
- 7.Hechinger RF, Lafferty KD. 2005. Host diversity begets parasite diversity: bird final hosts and trematodes in snail intermediate hosts. Proc. R. Soc. B 272, 1059–1066. ( 10.1098/rspb.2005.3070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thieltges DW, Hof C, Dehling DM, Brändle M, Brandl R, Poulin R. 2011. Host diversity and latitude drive trematode diversity patterns in the European freshwater fauna. Glob. Ecol. Biogeogr. 20, 675–682. ( 10.1111/j.1466-8238.2010.00631.x) [DOI] [Google Scholar]
- 9.Kamiya T, O'Dwyer K, Nakagawa S, Poulin R. 2014. Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography 37, 689–697. ( 10.1111/j.1600-0587.2013.00571.x) [DOI] [Google Scholar]
- 10.Cribb TH, Bott NJ, Bray RA, McNamara MKA, Miller TL, Nolan MJ, Cutmore SC. 2014. Trematodes of the Great Barrier Reef: emerging patterns of diversity and richness in coral reef fishes. Int. J. Parasitol. 44, 929–939. ( 10.1016/j.ijpara.2014.08.002) [DOI] [PubMed] [Google Scholar]
- 11.Poulin R, Presswell B. 2016. Taxonomic quality of species descriptions varies over time and with the number of authors, but unevenly among parasite taxa. Syst. Biol. 65, 1107–1116. ( 10.1093/sysbio/syw053) [DOI] [PubMed] [Google Scholar]
- 12.Martin LJ, Blossey B, Ellis E. 2012. Mapping where ecologists work: biases in the global distribution of terrestrial ecological observations. Front. Ecol. Environ. 10, 195–201. ( 10.1890/110154) [DOI] [Google Scholar]
- 13.Amano T, Sutherland WJ. 2013. Four barriers to the global understanding of biodiversity conservation: wealth, language, geographical location and security. Proc. R. Soc. B 280, 20122649 ( 10.1098/rspb.2012.2649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellard C, Jeschke JM. 2016. A spatial mismatch between invader impacts and research publications. Conserv. Biol. 30, 230–232. ( 10.1111/cobi.12611) [DOI] [PubMed] [Google Scholar]
- 15.Google Earth. 2017. Google Earth Version 7.3.0. Retrieved from https://www.google.com/earth/.
- 16.IUCN. 2017. The IUCN Red List of Threatened Species, Version 5.2. See http://www.iucnredlist.org.
- 17.BirdLife International and Handbook of the Birds of the World. 2016. Bird species distribution maps of the world. Version 6.0. See http://datazone.birdlife.org/species/requestdis.
- 18.Vilela B, Villalobos F. 2015. letsR: a new R package for data handling and analysis in macroecology. Methods Ecol. Evol. 6, 1229–1234. ( 10.1111/2041-210X.12401) [DOI] [Google Scholar]
- 19.Legendre P, Legendre L. 1998. Numerical ecology, second English edition Amsterdam, the Netherlands: Elsevier. [Google Scholar]
- 20.Zuur AF, Leno EN, Elphick CS. 2010. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 1, 3–14. ( 10.1111/j.2041-210X.2009.00001.x) [DOI] [Google Scholar]
- 21.Dormann CF, et al. 2007. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628. ( 10.1111/j.2007.0906-7590.05171.x) [DOI] [Google Scholar]
- 22.Clifford P, Richardson S, Hémon D. 1989. Assessing the significance of the correlation between two spatial processes. Biometrics 45, 123–144. ( 10.2307/2532039) [DOI] [PubMed] [Google Scholar]
- 23.Dutilleul P. 1993. Modifying the t test for assessing the correlation between two spatial processes. Biometrics 49, 305–312. ( 10.2307/2532625) [DOI] [Google Scholar]
- 24.Osorio F, Vallejos R.2014. SpatialPack: package for analysis of spatial data. R package version 0.2-3. http://cran.r-project.org/package=SpatialPack .
- 25.Fortin M-J. 1999. Effects of sampling unit resolution on the estimation of spatial autocorrelation. Ecoscience 6, 636–641. ( 10.1080/11956860.1999.11682547) [DOI] [Google Scholar]
- 26.Fortin M-J, Payette S. 2002. How to test the significance of the relation between spatially autocorrelated data at the landscape scale: a case study using fire and forest maps. Ecoscience 9, 213–218. ( 10.1080/11956860.2002.11682707) [DOI] [Google Scholar]
- 27.Moran PAP. 1950. Notes on continuous stochastic phenomena. Biometrika 37, 17–23. ( 10.2307/2332142) [DOI] [PubMed] [Google Scholar]
- 28.Tjøstheim D. 1978. A measure of association for spatial variables. Biometrika 56, 109–114. ( 10.2307/2335284) [DOI] [Google Scholar]
- 29.Matheron C. 1965. Les variables régionalisées et leur estimation. Paris, France: Masson. [Google Scholar]
- 30.Glick BJ. 1982. A spatial rank-order correlation measure. Geogr. Anal. 14, 177–181. ( 10.1111/j.1538-4632.1982.tb00066.x) [DOI] [Google Scholar]
- 31.Vallejos R. 2008. Assessing the association between two spatial or temporal sequences. J. Appl. Stat. 35, 1323–1343. ( 10.1080/02664760802382418) [DOI] [Google Scholar]
- 32.Vallejos R. 2012. Testing for the absence of correlation between two spatial or temporal sequences. Pattern Recogn. Lett. 33, 1741–1748. ( 10.1016/j.patrec.2012.05.013) [DOI] [Google Scholar]
- 33.R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 34.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451, 990–993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith KF. 2009. Global pathogen distributions: a win-win for disease ecology and biogeography. EcoHealth 6, 479–480. ( 10.1007/s10393-010-0304-3) [DOI] [PubMed] [Google Scholar]
- 36.Cribb TH, Bray RA, Diaz PE, Huston DC, Kudlai O, Martin SB, Yong RQY, Cutmore SC. 2016. Trematodes of fishes of the Indo-West Pacific: told and untold richness. Syst. Parasitol. 93, 237–247. ( 10.1007/s11230-016-9625-0) [DOI] [PubMed] [Google Scholar]
- 37.Costello MJ. 2016. Parasite rates of discovery, global species richness and host specificity. Integr. Comp. Biol. 56, 588–599. ( 10.1093/icb/icw084) [DOI] [PubMed] [Google Scholar]
- 38.Grenyer R, et al. 2006. Global distribution and conservation of rare and threatened vertebrates. Nature 444, 93–96. ( 10.1038/nature05237) [DOI] [PubMed] [Google Scholar]
- 39.Mecenero S, Altwegg R, Colville JF, Beale CM. 2015. Roles of spatial scale and rarity on the relationship between butterfly species richness and human density in South Africa. PLoS ONE 10, e0124327 ( 10.1371/journal.pone.0124327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulin R, Mouillot D. 2005. Host specificity and the probability of discovering species of helminth parasites. Parasitology 130, 709–715. ( 10.1017/S0031182004007218) [DOI] [PubMed] [Google Scholar]
- 41.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl Acad. Sci. USA 105, 11 482–11 489. ( 10.1073/pnas.0803232105) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Species distribution data are available in polygon format on the IUCN Red List of Threatened Species database (http://www.iucnredlist.org/technical-documents/spatial-data), and from BirdLife International (http://datazone.birdlife.org/species/requestdis). The full dataset on the geographical distribution of parasite species descriptions and the R script used in the analysis are uploaded as electronic supplementary material.