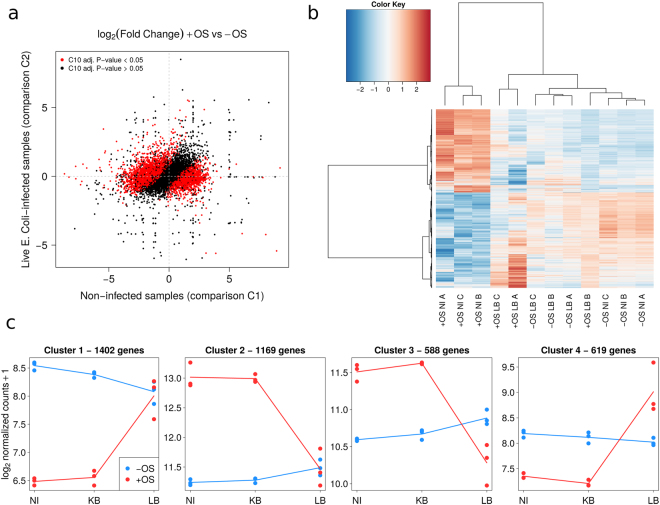
Figure 2.
Identification of genes whose expression was modified by incubation with E. coli O55 and exposure to OS. On the basis of the transcriptome data, a bioinformatics analysis identified genes under the influence of both OS and LB. (a) Illustration of the statistical interaction between OS and LB. A plot of the log2(FC) of the stress effect (+OS vs −OS) for LB samples (y-axis, comparison C2) vs. NI samples (x-axis, comparison C1) (Table S1). Red dots correspond to genes with a significant OS-bacterium interaction (comparison C10, n = 3778 with an adjusted p-value ≤ 0.05), i.e. genes with a log2(FC) for C1 that differed significantly from the log2(FC) for C2. (b) A heat map of gene expression values, showing the clustering within the C10 set of genes. Variance-stabilizing transformed counts were used to plot the heat map. The selected genes were associated with a statistically significant stress-bacterium interaction (adjusted p-value ≤ 0.05 for comparison C10, n = 3778). Row/genes values are centred on 0 but not scaled. The genes were hierarchically clustered using the correlation distance and the Ward aggregation criterion. (c) Average profiles for the four gene clusters defined by the hierarchical clusters shown in (b). The average transcriptome profile was established by computing the mean log2-normalized count for the genes in each of the four identified clusters.