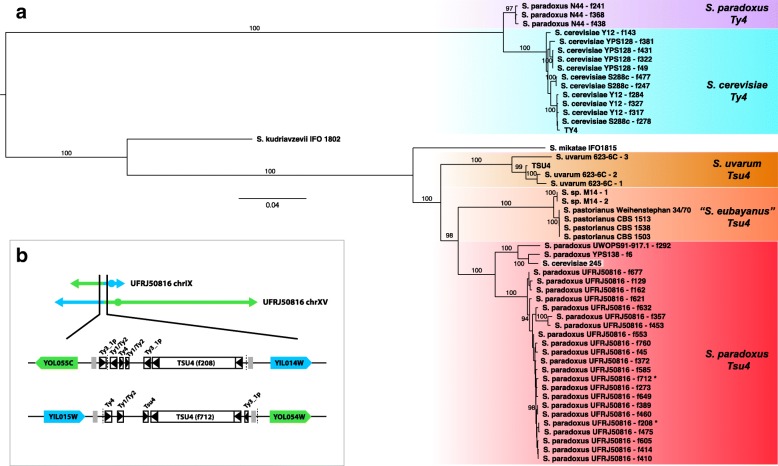
Fig. 1.
Evolution of the Ty4/Tsu4 super-family in the Saccharomyces sensu stricto species group. a. Maximum likelihood phylogeny of all full-length Ty4 and Tsu4 elements from 12 strains of S. cerevisiae and S. paradoxus with PacBio assemblies from Yue et al. [13] plus all complete or nearly-complete Tsu4 elements identified in Saccharomyces WGS assemblies at NCBI. Labels are shown for branches in the maximum likelihood tree that are supported by ≥ 90% of bootstrap replicates. The scale bar for branch lengths is in substitutions per site, and the tree is midpoint rooted. Tsu4 sequences from hybrid strains (S. pastorianus and Saccharomyces sp. M14) were assigned to the S. eubayanus subgenome and presumed to have originated in S. eubayanus. b. Genome organization of the reciprocal translocation between chrIX and chrXV in UFRJ50816. Sequences from the standard arrangement chrIX are shown in blue, and sequences from the standard arrangement chrXV are shown in green. Protein-coding genes, tRNA genes, and solo LTRs are shown approximately to scale as solid arrows, grey rectangles and boxed arrowheads, respectively. Approximate translocation breakpoints in UFRJ50816 based on whole genome alignments can be localized to chrIX:252268-259232 and chrXV:320536-328356 (dashed lines). Full-length Tsu4 elements are present in both translocation breakpoints. The Tsu4 elements associated in the chrIX and chrXV reciprocal translocation between are denoted by asterices in panel a