Figure 4.
JMJD1A/JMJD1B-Mediated H3K9 Demethylation Targets Gene-Dense Euchromatin
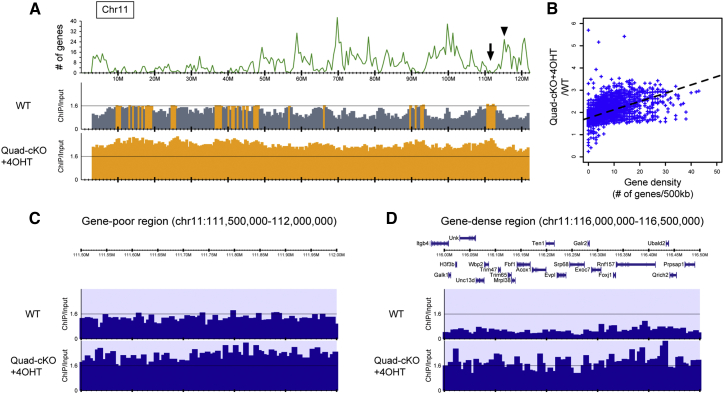
(A) Distribution profiles of H3K9me2 along chromosome 11. The upper panel shows the number (#) of mm10 RefSeq genes smoothed with a width of 500 kb. The middle and lower panels represent the ratios of normalized read density between ChIP and whole-cell lysate (input) samples (ChIP/input) in wild-type and JMJD1A/JMJD1B-depleted cells (Quad-cKO+4OHT), respectively. The ratio of ChIP/input >1.6 is shown in orange.
(B) Correlation between gene density and increased H3K9me2 levels due to JMJD1A/JMJD1B depletion. The x and y axes indicate the number of mm10 RefSeq genes and the quotient of ChIP/input of Quad-cKO+4OHT divided by ChIP/input of wild-type (WT), respectively, smoothed with a 500 kb width. The dashed lines represent regression lines.
(C and D) Distribution profile of H3K9me2 in a gene-poor region (C), arrow showing the position in (A), or a gene-dense region (D), arrowhead showing the position in (A) within chromosome 11. The upper panels show the positions of the protein-coding genes, whereas lower tracks depict ChIP/input in the indicated samples.