Figure 6.
Collaborative Roles of JMJD1A/JMJD1B Demethylases and G9A Methyltransferase for Tuning the H3K9 Methylation Levels and Regulating ESC Function
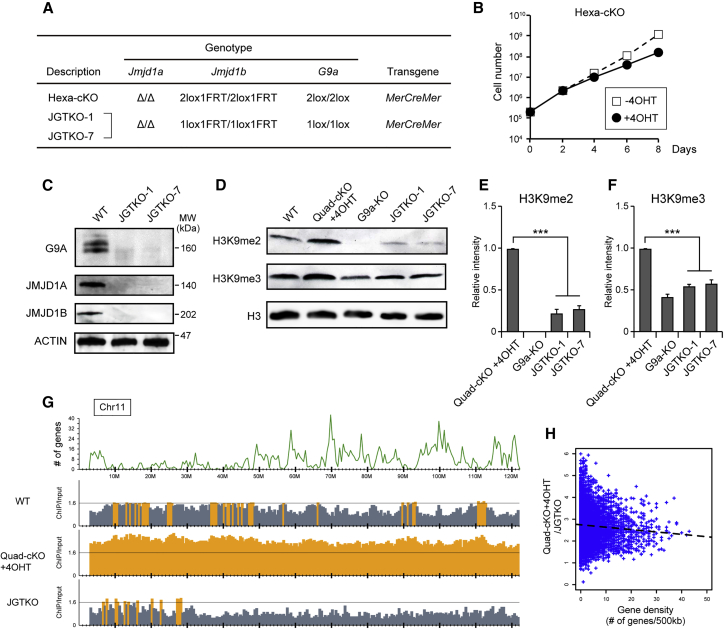
(A) Genotypes of the established ESC line. Hexa-cKO cell line carries alleles for the null mutation of Jmjd1a, the conditional mutation of Jmjd1b, and the conditional mutation of G9a. ESC lines lacking JMJD1A, JMJD1B, and G9A (named JGTKO-1 and JGTKO-7) were established by cloning from a pool of Hexa-cKO cells cultured in the presence of 4OHT.
(B) Growth curve of Hexa-cKO cell line in the presence or absence of 4OHT.
(C) JMJD1A, JMJD1B, and G9A proteins were absent in JGTKO-1 and JGTKO-7 ESC lines. Whole-cell extracts were fractionated by SDS-PAGE, and then subjected to immunoblot analysis with the indicated antibodies.
(D–F) G9a mutation rescues the H3K9 overmethylation phenotype of JMJD1A/JMJD1B-double-depleted cells. H3K9me2 and H3K9me3 levels in the indicated cell lines were examined by immunoblot analysis (D) and summarized (E and F, respectively). The intensities of H3K9me signals of Quad-cKO + 4OHT were defined as 1. Data are presented as means ± SD from n = 3 independent experiments. ∗∗∗p < 0.001 (Student's t test).
(G) Distribution profiles of H3K9me2 of JGTKO cells along chromosome 11. The top panel shows the number of mm10 RefSeq genes smoothed over 500 kb. The middle panels represent the ratios of normalized ChIP/input of ESCs of the indicated genotypes, including wild-type and JMJD1A/JMJD1B-depleted cells. The ratio of ChIP/input >1.6 is shown in orange.
(H) Correlation between gene density and decreased H3K9me2 levels due to G9a mutation in JMJD1A/JMJD1B-depleted cells. The x and y axes indicate the number of mm10 RefSeq genes and the quotient of ChIP/input of Quad-cKO+4OHT divided by ChIP/input of JGTKO cells, respectively, smoothed over 500 kb. Dashed lines represent regression curves.