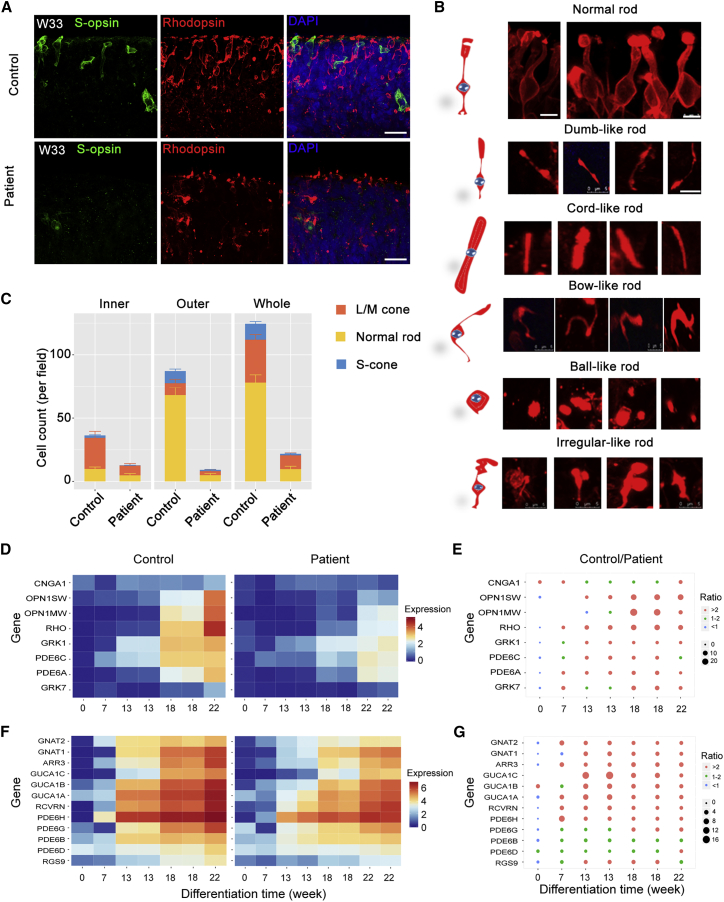
Figure 3.
Patient 3D Retinal Organoids Show Diseased Photoreceptors
(A) Immunostaining of the rod marker Rhodopsin (red) and the S-cone marker S-opsin (green) shows significant differences in cell morphology and cell counts in retinal organoids at W33. The same phenomenon has been observed in the L/M-cone (see also Figures S3G and S3I). Scale bar, 25 μm.
(B) Photoreceptor abnormalities can be observed in patient iPSC-derived 3D retina. Five subtypes of abnormal rod photoreceptors are identified based on the morphology. Scale bar, 5 μm.
(C) Comparison of photoreceptor counts in inner, outer, and whole-patient and control retinal organoids. Orange, yellow, and blue represent L/M-cones, normal rods, S-cones respectively. n = 3 organoids for each cell type. Data are from three independent experiments.
(D and F) Heatmaps illustrate the gene expression profile of retina-related genes at different organoid stages containing RNA-seq dataset at week 0, 7, 13, 18, and 22. Most of the genes exhibited elevated expression in control retinae compared with that of the patient from week 7. Different colors represent the value of In (FPKM [fragments per kilobase of transcript per million mapped reads] + 1).
(E and G) The size of the dot represents the FPKM ratio of control to patient. The genes correspond with those in D and F. Significant differences can be observed in photoreceptor-associated genes. Orange dots, >2 fold change; green dots, 1–2 fold change; blue dots, <1 fold change.