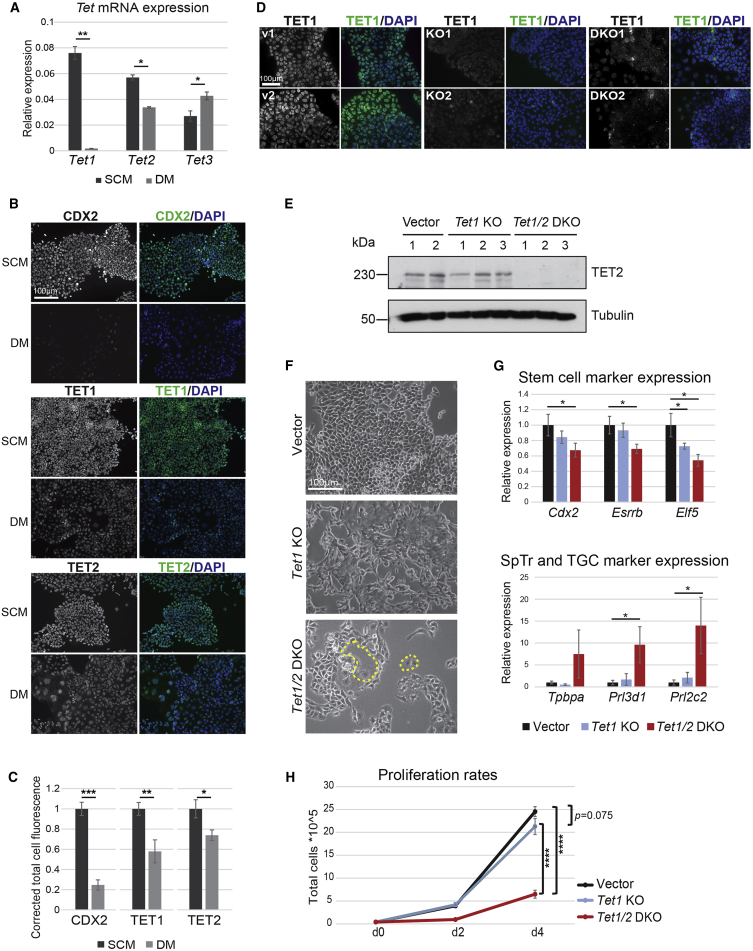
Figure 1.
TET1 and TET2 Positively Correlate with the TSC State
(A) qRT-PCR analysis of Tet1,2,3 mRNA expression in TSCs cultured in stem cell media (SCM) or differentiation media (DM) over 3 days. Data are normalized against housekeeping genes Sdha and Dynein, and are displayed as mean ± SEM; ∗p < 0.05, ∗∗p < 0.01 (ANOVA with Holm-Bonferroni post-hoc test); n = 3 independent replicates each.
(B) Immunofluorescence (IF) staining for CDX2, TET1, and TET2 in TSCs cultured in SCM and DM. Scale bar, 100 μm.
(C) Quantification of total mean cell fluorescence of (B). ∗p < 0.05, ∗∗p < 0.01; ∗∗∗p < 0.001 (unpaired, one-sided t test). Measurements are of ≥100 cells each.
(D) IF staining for TET1 in independent vector (v) control and Tet1 KO and Tet1/2 DKO clones. Scale bar, 100 μm.
(E) Western blot for TET2 on vector control, Tet1 KO and Tet1/2 DKO TSCs. Tubulin was used as loading control.
(F) Phase contrast images of vector control, Tet1 KO and Tet1/2 DKO clones grown in SCM. Yellow dotted lines indicate enlarged giant cell-like cells. Scale bar, 100 μm.
(G) qRT-PCR analysis of TSCs and differentiation markers in vector control (set to 1) and mutant clones. SpTr, spongiotrophoblast; TGC, trophoblast giant cells. Data are mean ± SEM; ∗p < 0.05 (ANOVA with Holm-Bonferroni post-hoc test); n = 4 clones as independent replicates each.
(H) Analysis of proliferation rates over a 4-day period. ∗∗∗∗p < 0.0001 (two-way ANOVA with Holm-Sidak's multiple comparisons test); n = 5 independent replicates each.
See also Figures S1–S3.