Figure 2.
Tet1 and Tet1/2 Depletion Induces EMT and Centrosome Defects
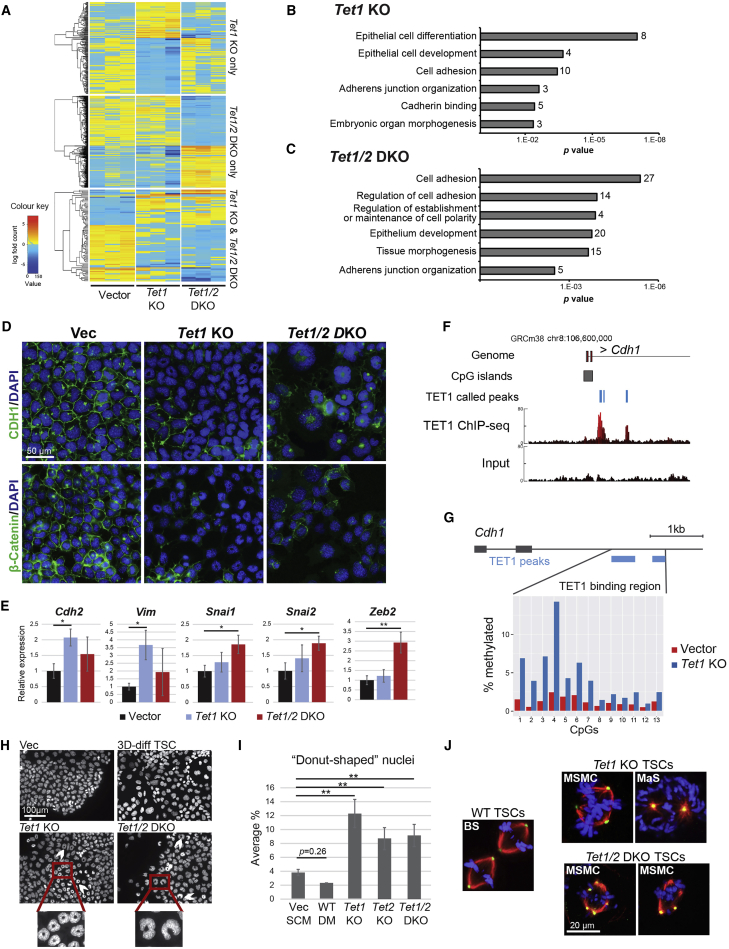
(A) Heatmap of differentially expressed genes between vector, Tet1 KO, and Tet1/2 DKO clones. n = 3 independent clones each.
(B and C) Gene ontology term enrichment analysis for differentially expressed genes in Tet1 KO cells (B) and in Tet1/2 DKO cells (C). The numbers of genes in each enrichment term are also given.
(D) IF staining for E-cadherin (CDH1) and β-catenin. Scale bar, 50 μm.
(E) qRT-PCR expression analysis of mesenchymal markers. Data are mean ± SEM; ∗p < 0.05, ∗∗p < 0.01 (ANOVA with Holm-Bonferroni post-hoc test); n = 4 clones as independent replicates each.
(F) TET1 ChIP-seq data displayed on a genome browser view for the Cdh1 locus. Called peaks and actual peaks are displayed.
(G) Bisulfite sequencing analysis across the TET1-bound region of the Cdh1 locus, encompassing 13 individual CpG's, on vector control and Tet1 KO TSCs.
(H) DAPI-stained nuclei of vector, Tet1 KO, and Tet1/2 DKO clones grown in SCM, and 3-day differentiated WT TSCs. The white arrows point at ring- (donut) or crescent-shaped nuclei. Scale bar, 100 μm.
(I) Unbiased quantification of donut-shaped nuclei by ImageJ analysis. WT DM are 3-day differentiated WT TSCs as shown in (H). ∗∗p < 0.01 (two-way ANOVA with Bonferroni post-hoc test). Measurements are of >400 cells each.
(J) IF staining for α-tubulin (red), γ-tubulin (green), and chromatin (blue) in WT TSCs, Tet1 KO, and Tet1/2 DKO TSCs. Confocal images of representative spindle structures, including bipolar spindles (BS) in WT TSCs, multiastral spindles with multiple centrosomes (MSMC) resulting in misaligned chromosomes, and monoastral spindles (MaS), are shown. Scale bar, 20 μm.
See also Figure S4.