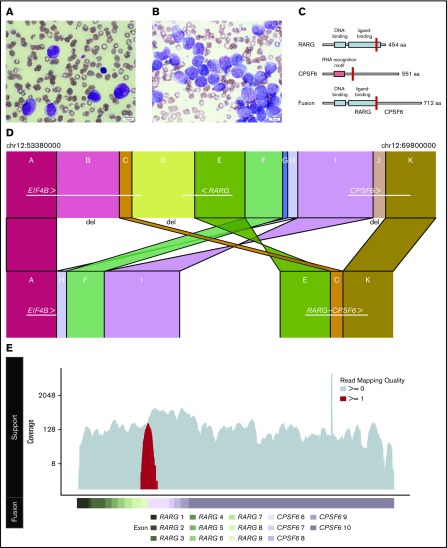
Figure 1.
Histopathologic and genomic characterization of a case of AML with promyelocytic features. (A) Wright-Giemsa staining of the peripheral blood smear highlighting promyelocytes (original magnification ×100). (B) Wright-Giemsa staining of the aspirate from the diagnostic bone marrow biopsy highlighting numerous blasts and promyelocytes (original magnification ×100). The images were captured by an Olympus BX53 microscope with an Olympus UPlanSApo 100×/1.4 oil objective and an Olympus DP26 digital camera with Olympus cellSens standard software (version 1.16; Tokyo, Japan). (C) Representative schematic of the main protein domains of RARG isoform 1 (top), CSPF6 isoform 2 (middle), and the predicted RARG-CPSF6 fusion (bottom) with the fusion breakpoints highlighted in red (for RARG at amino acid 392 and for CPSF6 at amino acid 231). Isoform choice was based on read support from the RNA-Seq data. (D) Schematic of the highly rearranged region on chromosome 12 where the reciprocal inversion occurred. In panel D, regions A and K bookend regions B-J that were involved in distinct deletions or rearrangement events. The segments are not to scale. (E) RNA-Seq reads based on a pseudo-alignment to the RARG-CPSF6 predicted fusion transcript. The coverage of reads uniquely aligned to the predicted fusion (spanning the fusion breakpoint) is displayed in red on the top track in relation to the exons for the predicted fusion (bottom track). We observed only three reads supporting wild-type RARG expression, which are likely indicative of a small number of contaminating benign cells.