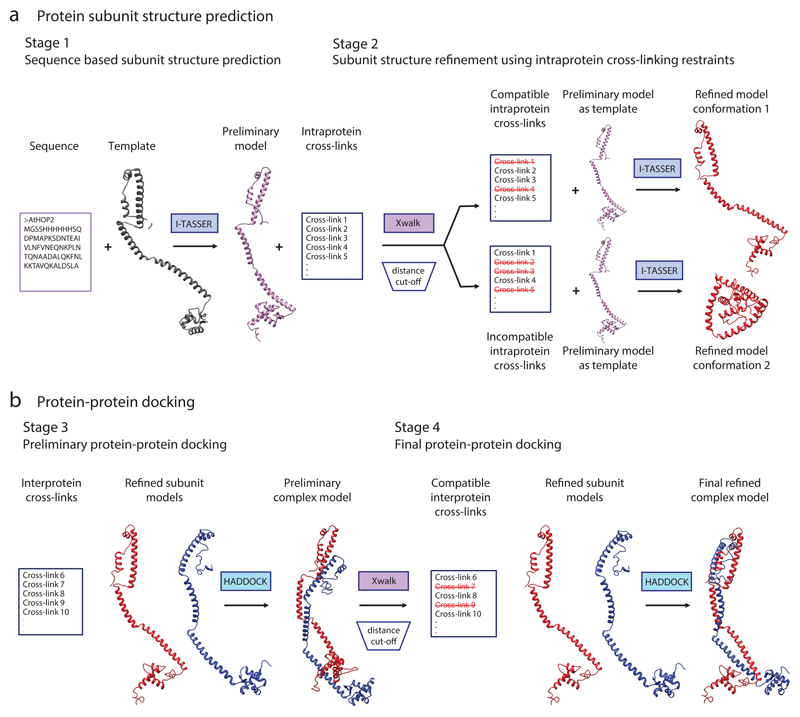
Figure 1. Overview of the structural modeling workflow using XL-MS data.
(a) Schematic illustration of an example modeling procedure for protein subunit structure. Stage 1: a preliminary model of the protein is generated using the I-TASSER algorithm. Here, the structure is predicted on the basis of the primary amino acid sequence and the structure of a homologous template. Stage 2: the distances between the residues of the intraprotein cross-links are calculated within the preliminary model with Xwalk. The cross-links are subsequently divided according to their compatibility with the preliminary structure; those that bridge residues that are close together in 3D space are compatible with the model and are used to generate refined model 1. The cross-links that do not satisfy the model can be used to predict an alternative conformation (conformation 2, Stage 2). In this case, refined model conformation 1 corresponds to an open conformation, whereas refined model conformation 2 reflects a closed conformation. (b) Schematic illustration of the docking procedure for a protein dimer. Stage 3: a preliminary complex model is generated using the HADDOCK protein–protein docking tool, which combines the subunit structures obtained from Stage 2 with interprotein cross-link data. Stage 4: interprotein cross-links are divided according to their compatibility with the preliminary complex model. Compatible cross-links are used for a second round of protein complex docking with HADDOCK, generating the final refined protein complex model.