Figure 2.
Dynamic Localization of KLF4 Alters Nuclear Complexes during Exit from Naive Pluripotency
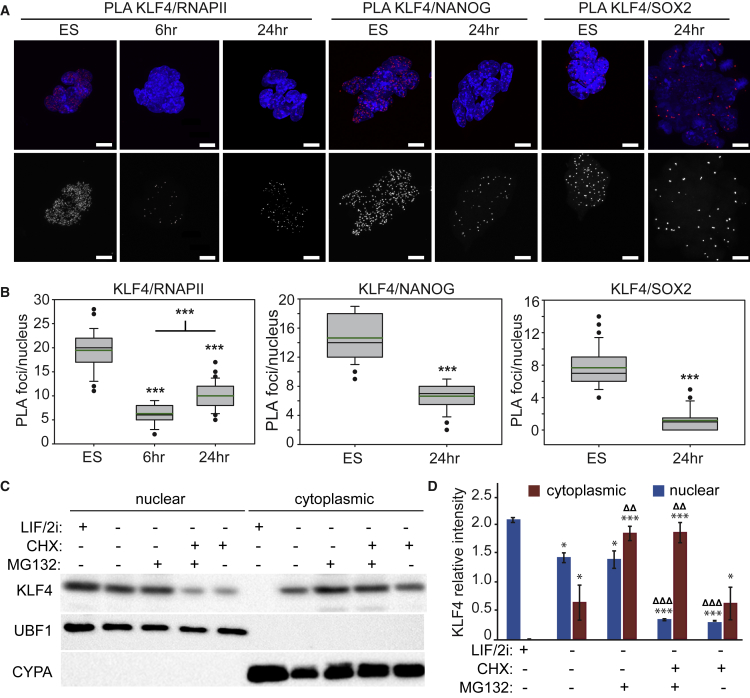
(A) Proximity ligation amplification (PLA) indicating the interaction between KLF4/RNAPII-S5P, KLF4/NANOG, and KLF4/SOX2 in ESCs are disrupted 6 or 24 hr after LIF/2i withdrawal. Images shown are maximum-intensity projections. Scale bars, 10 μm.
(B) Box-and-whisker plots indicate the number of PLA foci per nucleus. Boxes indicate interquartile range of intensity values and whiskers indicate the 10th and 90th percentiles; outliers are shown as black dots. Images were collected from at least three biological replicate samples and ≥100 nuclei were quantified for each. Statistical differences are indicated by ∗∗∗p < 0.001.
(C) Immunoblot of nuclear and cytoplasmic fractions from ESCs after 24 hr LIF/2i withdrawal and 12 hr treatment with CHX or MG132.
(D) Quantification of relative intensity levels of KLF4 in immunoblots from three biological replicates reveals that the proteasome inhibitor MG132 prevents degradation of cytoplasmic KLF4 while the protein synthesis inhibitor cycloheximide (CHX) prevents accumulation of nuclear KLF4. Statistical differences compared with 0-hr values are indicated by ∗p < 0.05 and ∗∗∗p < 0.001; statistical differences between untreated 24-hr differentiated fractions (LIF/2i−) and CHX or MG132 treated fractions are indicated by ΔΔp < 0.01 and ΔΔΔp < 0.001. Error bars represent standard deviation. See also Figure S2.