Figure 5.
The KLF4 ERK Phosphorylation Site S132, NES1, and NES2 Are Required for KLF4 Nuclear Export
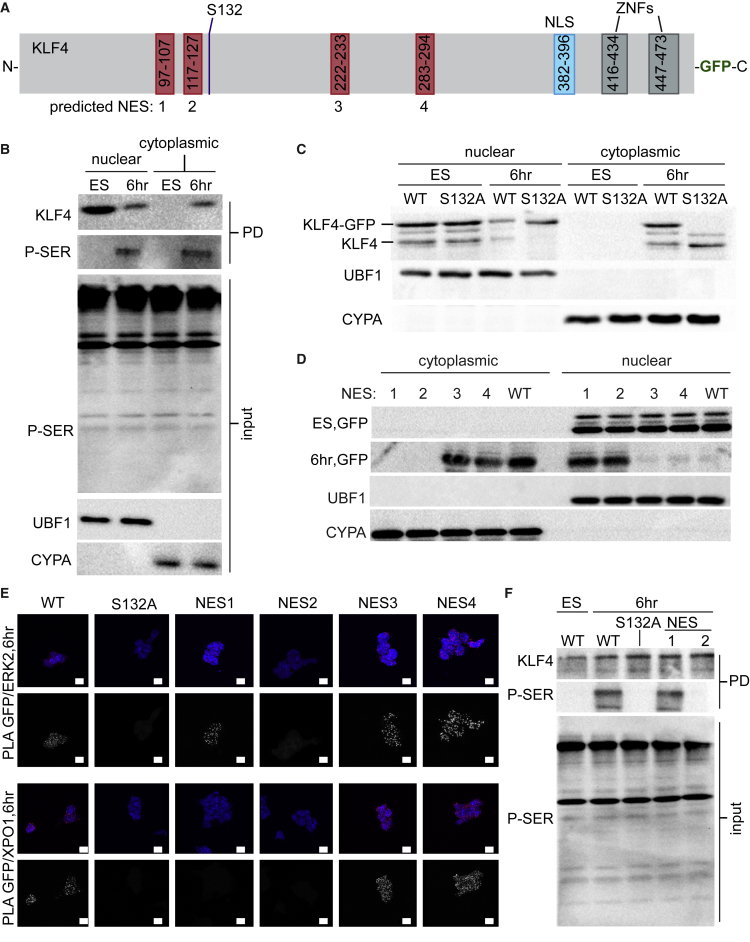
(A) Schematic of mouse KLF4 depicting predicted nuclear export signals (NES), nuclear localization signal (NLS), ERK phosphorylation site S132, and zinc fingers (ZNFs).
(B) Immunoprecipitated KLF4 protein from nuclear and cytoplasmic fractions of ESCs cultured with LIF/2i and 6 hr after LIF/2i removal, probed with anti-KLF4 and anti-phosphoserine (P-SER; PD, pull-down). Cyclophilin A (CYPA) and the nucleolar protein upstream binding factor (UBF1) reveal purity of the cytoplasmic and nuclear fractions, respectively.
(C) Nuclear and cytoplasmic fractions prepared from KLF4-GFP ES lines indicate that wild-type (WT) KLF4-GFP (top band) but not KLF4S132A-GFP is exported to the cytoplasm 6 hr after LIF/2i removal. Endogenous KLF4 (bottom band) is exported to the cytoplasm in both cases.
(D) Nuclear and cytoplasmic fractions prepared from WT KLF4-GFP and KLF4-GFP NES mutants (1–4). Anti-GFP immunoblot indicated that NES1 and NES2 are required for nuclear export of KLF4 after removal of LIF/2i.
(E) Proximity ligation amplification (PLA) for GFP/ERK2 and GFP/XPO1 indicated that the interaction between KLF4/ERK2 was disrupted by S132 and NES2 mutations. The interaction between KLF4/XPO1 was disrupted by S132, NES1, and NES2 mutations. Images shown are maximum-intensity projections. Scale bars, 10 μm.
(F) Anti-phosphoserine immunoblot of GFP-Trap revealed that S132A and NES2 mutations disrupted KLF4 serine phosphorylation.
See also Figure S5.