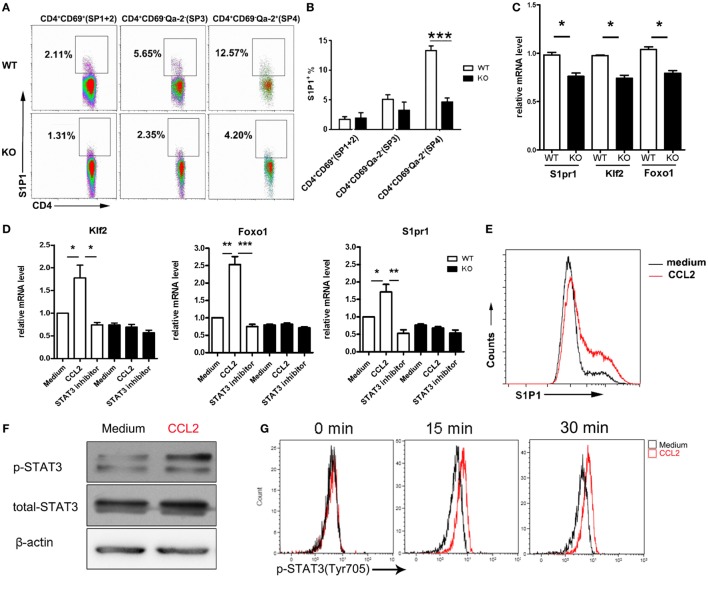
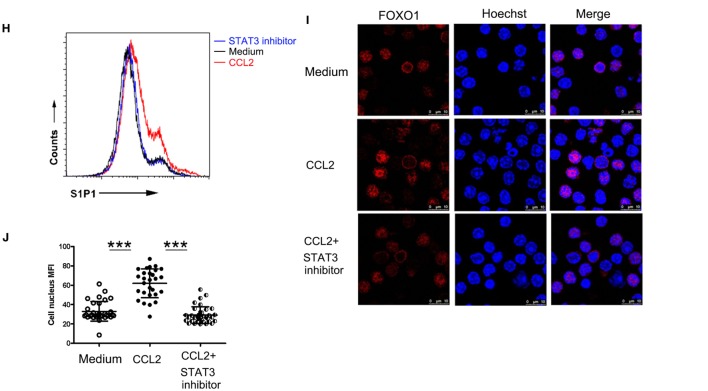
Figure 5.
Activation of the FoxO1–KLF2–S1P1 axis by CCR2 signaling through Stat3. (A) Surface expression of S1P1 in the CD69+Qa-2− (SP1 + SP2), CD69−Qa-2− (SP3), and CD69−Qa-2+ (SP4) subsets of CD4 SP thymocytes from WT and KO mice. The experiments were repeated three times with six mice for each group. Representative dot plots are shown. (B) Percentage of S1P1+ cells are presented as mean ± SD. (C) Foxo1, Klf2, and S1pr1 mRNA expression in WT and KO CD4 SP thymocytes as determined by qPCR. The experiments were repeated three times with triplicates. Data are present as mean ± SEM. (D) CD69−Qa-2+ SP4 thymocytes from WT and KO mice were stimulated with CCL2 in the presence or absence of STAT3 inhibitor Stattic. Foxo1, Klf2, and S1pr1 mRNA expression was detected by qPCR. The experiments were repeated three times with triplicates. Data are present as mean ± SEM. (E) Following stimulation with CCL2, WT SP4 thymocytes were examined for surface expression of S1P1. Representative histograms are shown out of three independent experiments. (F,G) Levels of phosphorylated Stat3 were examined by Western blotting (F) or intracellular staining (G) in SP4 thymocytes after stimulation with CCL2. Western blotting was performed at 30 min after stimulation. The experiments were repeated three times with similar results. (H–J) SP4 thymocytes were stimulated with CCL2 in the presence or absence of STAT3 inhibitor Stattic. Surface expression of S1P1 was assayed by flow cytometry (H). Subcellular localization of FoxO1 was revealed by immunofluorescent staining (I) and nucleus Foxo1 was quantified by mean fluorescent intensity (MFI) (J). Data are presented as mean ± SD. Two independent experiments were performed with similar results. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: ns, no significance; S1P1, sphingosine-1-phosphate receptor 1; SP, single-positive; WT, wild-type; KO, knockout; KLF2, Kruppel-like factor 2; FoxO1, forkhead box O1.