Figure 1.
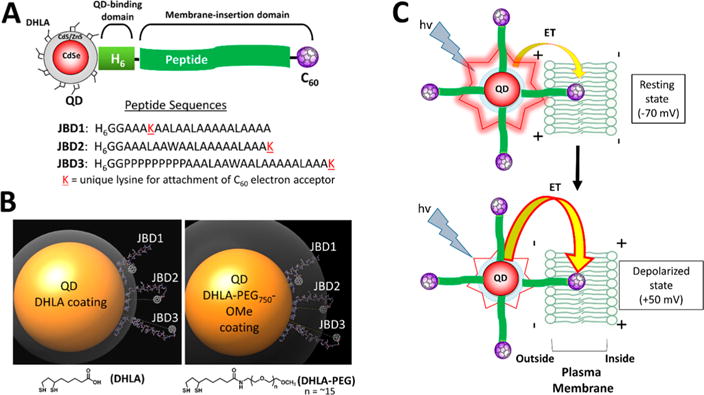
QD–peptide–C60 bioconjugates. (A) Modular design of QD–peptide–C60 bioconjugates. The core/shell QD serves as a central scaffold to which is appended peptides bearing a C60 fullerene at discrete positions. A unique lysine (K) allows for the controlled placement of the C60 moiety at different distances from the QD surface. An N-terminal polyhistidine tract (H6) mediates self-assembly of the peptide to the QD shell. Peptide sequences are written in N → C orientation. (B) Molecular models of QD–peptide–C60 bioconjugates. Shown is the 605 nm emitting core/shell QD (yellow sphere; 100 Å in diameter) capped with DHLA (left panel) and DHLA–PEG750–OMe (right panel) ligands for water solubility. The ligand layer is shown in gray and is 11 Å (DHLA) or 33 Å thick (DHLA–PEG750–OMe). Each QD is shown appended with peptides JBD1, JBD2, and JBD3 wherein the C60 electron acceptor is iteratively positioned at increasingly further distances from the QD surface. The predicted surface-to-surface distances from the QD to the C60 surface are as follows: JBD1, 10 Å; JBD2, 24 Å; JBD3, 42 Å. (C) Schematic of the response of QD–peptide–C60 bioconjugates to changes in membrane potential. Multiple peptide–C60 conjugates are arrayed around the central QD (electron donor), and they all engage in electron transfer (ET) with the QD; a percentage of the arrayed peptide–C60 are inserted into the membrane bilayer and contribute to depolarization-induced PL quenching. At resting potential, minimal ET from the photoexcited QD to those C60 embedded in the membrane bilayer results in bright QD emission (top). Depolarization of the membrane potential augments the rate of ET, causing a decrease in QD PL (bottom).
