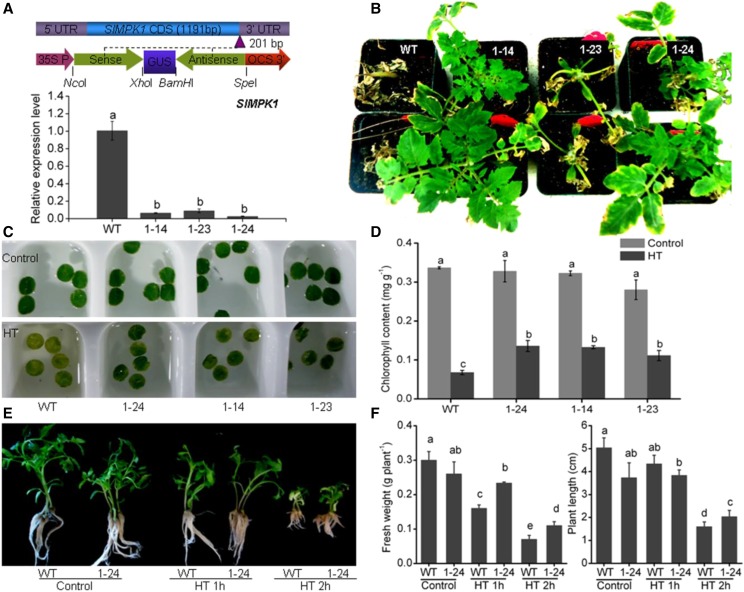
Figure 2.
Effects of SlMPK1 suppression on HT tolerance of tomato seedlings. A, Schematic diagram of the RNAi-SlMPK1 construct and the expression level of SlMPK1 in RNAi-SlMPK1 plants. The closed green arrows represent the partial sequence of SlMPK1 (top). UTR, Untranslated regions; CDS, coding sequence; 35S P, 35S promoter; OCS, OCS terminators. The restriction sites used are shown. The graph at bottom shows reverse transcription-quantitative PCR (RT-qPCR) analysis of the transcript levels of SlMPK1 of partial transgenic lines. The numbers indicate independent RNAi transgenic plants. B, Phenotypes of RNAi-SlMPK1 lines in nutritional bowl feeding matrix soils under HT. Seedlings of the wild type (WT) and RNAi-SlMPK1 lines 1-14, 1-23, and 1-24 at the five-leaf stage were subjected to 38°C/28°C for 3 d and then recovered at 25°C/20°C for 10 d. C, Phenotypes of leaf discs in RNAi-SlMPK1 lines under HT. The leaf discs from fourth fully expanded leaves were incubated at 45°C for 2 h, followed by recovery at 25°C/20°C for 2 d. D, Chlorophyll content of the leaf discs of RNAi-SlMPK1 transgenic plants under HT shown in C. Error bars represent sd values of three replicates. E, Phenotypes of RNAi-SlMPK1 line 1-24 in Murashige and Skoog medium under HT. F, Fresh weight and plant length of the wild-type and 1-24 plants shown in E. Statistical differences among the samples shown in A, D, and F are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA).