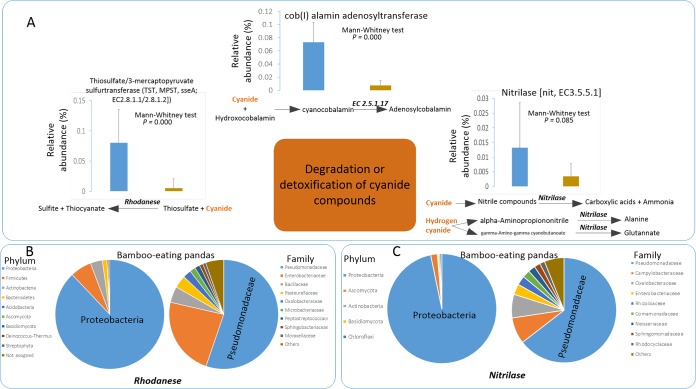
FIG 3 .
Potential for cyanide compound degradation and detoxification by gut microbes from bamboo-eating pandas (giant pandas and red pandas). (A) The proportions of genes coding for the following putative vital enzymes that are related to the potential degradation and detoxification of cyanide compounds in gut microbiomes of bamboo-eating pandas: nitrilase (Nit) (EC 3.5.5.1), thiosulfate/3-mercaptopyruvate sulfurtransferase (encoded by TST gene, MPST gene, and sseA) (EC 2.8.1.1/2.8.1.2), and cob(I)alamin adenosyltransferase (EC 2.5.1.17). Blue bars, bamboo-eating pandas; dark yellow bars, Père David’s deer. (B) The taxonomic assignments of the identified genes (glpE, TST gene, MPST gene, and sseA) coding for thiosulfate/3-mercaptopyruvate sulfurtransferases (EC 2.8.1.1/2.8.1.2). (C) The taxonomic assignments of the recognized genes coding for nitrilase (EC 3.5.5.1). The relative abundances in panels B and C are the proportions of reads assigned to specific taxa in comparison to total reads (glpE, TST gene, MPST gene, and sseA) coding for putative thiosulfate/3-mercaptopyruvate sulfurtransferases (EC 2.8.1.1/2.8.1.2) or nitrilase (EC 3.5.5.1).