FIG 4 .
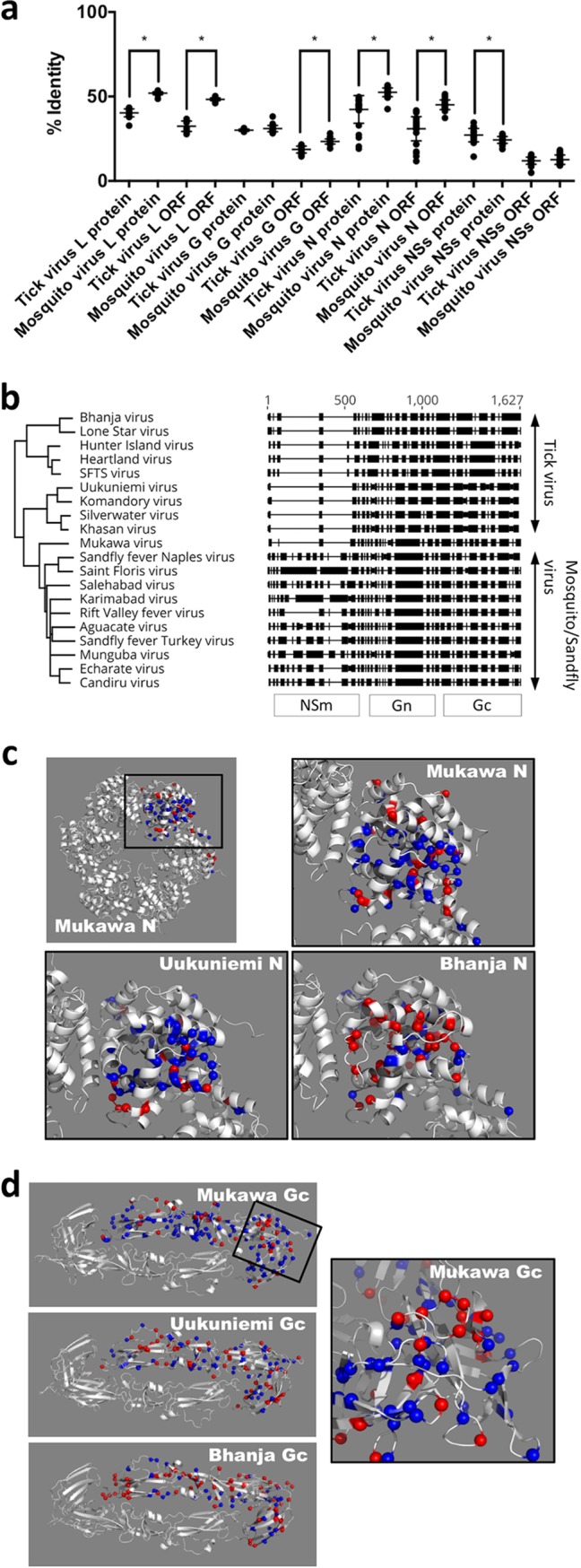
Molecular characterization of MKWV. (a) Nucleotide sequence identities of open reading frames or deduced amino acid sequence identities of viral proteins (RNA-dependent RNA polymerase [L], glycoprotein [G], nucleoprotein [N], or nonstructural protein [NSs]) were determined for the comparisons of MKWV with tick-borne phleboviruses (tick virus) and mosquito/sandfly-borne phleboviruses (mosquito virus). The mean identity of each group is presented as a horizontal bar. Significant differences between the identity values of tick virus comparisons and mosquito virus comparisons are indicated by asterisks (P < 0.05). (b) Alignments of deduced amino acid sequences of phlebovirus glycoproteins. Boxes indicate aligned amino acids, and bars indicate gaps in each sequence. (c and d) Structures of N protein (c) and membrane glycoprotein Gc (d) of three tick-borne phleboviruses, MKWV, Bhanja virus, and Uukuniemi virus, were predicted based on the crystal structures of RVFV proteins. Amino acid residues identical to those of RVFV and SFTSV are shown in blue and red, respectively. Amino acid residues shared with both RVFV and SFTSV are shown in dark gray. The inset view of the surface loop of domain II in the MKWV Gc is indicated by a black square.
