Abstract
Objective
The aim was to assess whether loci associated with metabolic traits also have a significant role in BMI and mental traits/disorders
Methods
We first assessed the number of single nucleotide polymorphisms (SNPs) with genome-wide significance for human metabolism (NHGRI-EBI Catalog). These 516 SNPs (216 independent loci) were looked-up in genome-wide association studies for association with body mass index (BMI) and the mental traits/disorders educational attainment, neuroticism, schizophrenia, well-being, anxiety, depressive symptoms, major depressive disorder, autism-spectrum disorder, attention-deficit/hyperactivity disorder, Alzheimer's disease, bipolar disorder, aggressive behavior, and internalizing problems. A strict significance threshold of p < 6.92 × 10−6 was based on the correction for 516 SNPs and all 14 phenotypes, a second less conservative threshold (p < 9.69 × 10−5) on the correction for the 516 SNPs only.
Results
19 SNPs located in nine independent loci revealed p-values < 6.92 × 10−6; the less strict criterion was met by 41 SNPs in 24 independent loci. BMI and schizophrenia showed the most pronounced genetic overlap with human metabolism with three loci each meeting the strict significance threshold. Overall, genetic variation associated with estimated glomerular filtration rate showed up frequently; single metabolite SNPs were associated with more than one phenotype. Replications in independent samples were obtained for BMI and educational attainment.
Conclusions
Approximately 5–10% of the regions involved in the regulation of blood/urine metabolite levels seem to also play a role in BMI and mental traits/disorders and related phenotypes. If validated in metabolomic studies of the respective phenotypes, the associated blood/urine metabolites may enable novel preventive and therapeutic strategies.
Keywords: Cross-trait analysis, Metabolites, Obesity, Schizophrenia, Intelligence, Educational attainment
Highlights
-
•
5–10% of genomic regions that regulate blood/urine metabolite levels were associated with BMI and mental phenotypes.
-
•
Metabolite loci were particularly associated with BMI, Alzheimer's disease, schizophrenia, and educational attainment.
-
•
Confirmation was achieved for BMI and educational attainment.
1. Introduction
Genome-wide association studies (GWAS) and meta-analyses thereof (GWAMA) have offered insights into the genetic makeup of BMI and of several categorically defined mental disorders [1] as well as dimensional mental phenotypes [2], [3], [4], [5]. The largest number of genome-wide significant loci have been identified for BMI, for which in total 209 loci have been detected in subjects of European and Japanese ancestry, respectively [6], [7]. Schizophrenia ranks highest among mental traits and disorders with currently 108 identified independent genetic loci (Supplemental Table 1); for other disorders/traits such as attention-deficit/hyperactivity disorder (ADHD) and internalizing traits, genome-wide significant loci have yet to be reported. Currently, only a small fraction of the respective heritability estimates can be explained at the DNA level and functional implications of the detected genetic variations have only insufficiently been elucidated.
A substantial amount of evidence links obesity with mental health [8]; obviously, behavior figures prominently in both energy intake and expenditure. Genetic factors seemingly account for part of this link. Thus, a number of lookups of SNP hits for one trait (e.g. BMI) in GWAMA data for a related trait (e.g. anorexia nervosa [9]) and vice versa have detected jointly relevant genetic variants. Such cross-phenotype/-trait analyses have led to the identification of potentially relevant genetic loci, which had not yet achieved genome-wide significance in the respective GWAMA analyses. Additionally, the identification of genetic correlations between BMI and mental traits/disorders suggests causal relationships [1], [10]. Thus, cross-trait linkage disequilibrium score regression (LDSC) revealed genetic correlations between BMI/obesity and schizophrenia, bipolar illness, anorexia nervosa, Alzheimer's disease, smoking behavior, neuroticism, and educational attainment [1]. Furthermore, genetic correlations have also been identified between different mental traits/disorders [11]; e.g. between bipolar disorder (BD) and schizophrenia [1].
In contrast to the weak effect sizes for loci identified in GWAMA of both BMI and mental phenotypes, GWAS performed for levels of blood or urine metabolites and ratios thereof have identified several loci with substantially larger effect sizes (Supplemental Table 2). For example, in the initial GWAS of 163 serum metabolic traits, which was based on only 1,809 population-based German probands (discovery) and 422 twins of a UK twin cohort (confirmation), the locus with the strongest effect size explained 36.3% of the ratio of two serum metabolites [12]. In addition, in the currently most comprehensive study, which reported 145 genome-wide significant independent SNP associations with blood metabolites, the median for the contribution of genome-wide significant metabolic loci to metabolite variance has been estimated at 6.9% (range 1–62%; [13]). Such loci may improve our understanding of genetic disease predisposition and enable identification of potential biomarkers, drug targets and the causal role of environmental and modifiable determinants in human traits and disease [13].
Metabolism related studies have been conducted for both BMI and mental disorders. For example, genetic factors in metabolism have been identified in deeply phenotyped mouse models [14]. Levels of key metabolites, e.g. branched chain amino acids, which are part of the cross-talk with lipid metabolism, have been evaluated in obesity [15] in humans. Metabolites have also been associated with mental phenotypes: In a small sample of 17 drug-free schizophrenia patients, plasma creatine levels were lower, while 2-hydroxybutyric acid levels were higher compared with 19 healthy controls [16]. In contrast, reduced betaine levels, which were previously suggested as a biomarker candidate at least for first-onset schizophrenia [17], were not replicated in this study. These conflicting results could at least partly be explained by differences in durations of illness between study samples since a positive correlation was observed between duration of illness and betaine concentrations [16]. Despite this research, a generally approved biomarker (panel) for schizophrenia still remains to be identified [18].
SNPs explaining a proportion of both the variance of specific metabolite concentrations and BMI and mental phenotypes could help identify candidate metabolites to be measured in serum samples of patients in order to determine if indeed the metabolite concentrations are directly associated. Knowledge of such associations may contribute to the development of new options for treatment and prevention of obesity and mental disorders via environmentally induced alterations of the respective metabolite levels [12], [19].
The aim of the current study was to assess whether loci associated with metabolic traits also have a significant role in the partially behavior driven phenotype BMI, the mental disorders schizophrenia, major depressive disorder (MDD), BD, autism-spectrum disorder (ASD), ADHD, and Alzheimer's disease, and the quantitative mental/behavioral phenotypes educational attainment, neuroticism, well-being, aggressive behavior, anxiety, depressive symptoms, and internalizing problems (all 14 phenotypes subsequently referred to as mental phenotypes). To that end, we performed a lookup analysis of SNPs with genome-wide significant association to metabolic traits in GWAS of these mental phenotypes. Finally, SNPs identified as being potentially relevant for BMI or educational attainment were looked up in independent data sets for confirmation.
2. Material and methods
2.1. Selection of relevant SNPs
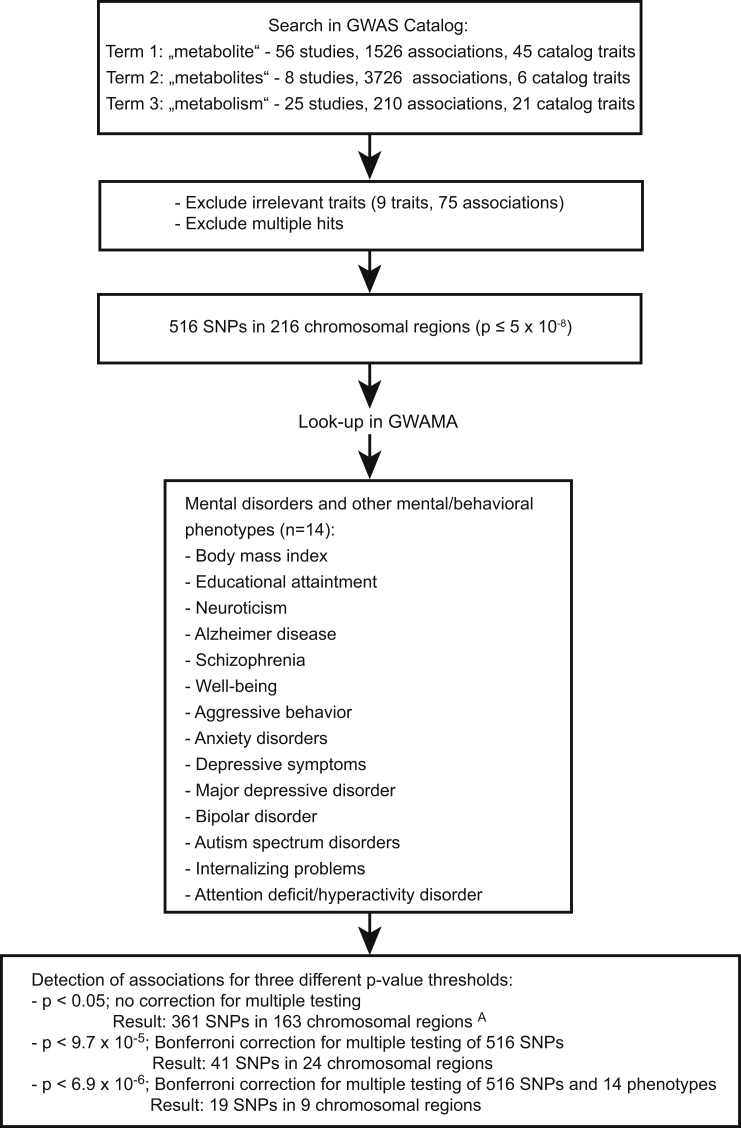
We performed a lookup of SNPs, which are genome-wide significantly associated with metabolite levels or ratios thereof (p ≤ 5 × 10−8, Figure 1). Relevant SNPs were derived from the GWAS Catalog [20] (The NHGRI-EBI Catalog of published genome-wide association studies: http://www.ebi.ac.uk/gwas/, accessed on December 12th 2016, version v1.0.) using the search terms ‘metabolite,’ ‘metabolites,’ and ‘metabolism’ (Figure 1). The resulting studies pertaining to metabolic traits were independently reviewed by two authors (JH, LL) in order to find traits, which are not directly related to physiological metabolism. Single different ratings were resolved by consensus. This selection process resulted in the exclusion of the following traits from further analyses: birth weight, cardiovascular heart disease in diabetics, response to serotonin reuptake inhibitor, hereditary hemochromatosis-related traits, AR-C124910XX levels in individuals with acute coronary syndromes treated with ticagrelor, metabolic traits in smokers, and nicotine metabolite ratio.
Figure 1.
Flow chart of the conducted lookup analysis. Legend: A30 associations were identified via proxy-SNPs; GWAMA: Genome-wide association meta-analysis.
The lookup of the identified SNPs was performed in the following publically available GWAMA data sets from: (a) the Psychiatric Genetics Consortium (PGC): all GWAMA data available until February 12th 2017 including schizophrenia [21], MDD [22], ADHD [23], BD [24] and ASD (downloaded between November 22nd 2016 and February 12th 2017) [25]; (b) the Social Science Genetic Association Consortium (SSGAC): educational attainment [3], neuroticism [2], well-being [2], depressive symptoms [2] (downloaded between November 22nd and December 12th 2016); (c) the Genetic Investigation of Anthropometric Traits (GIANT) Consortium: BMI [7] (downloaded on March 26th 2016); (d) Early Genetics and Lifecourse Epidemiology Consortium (EAGLE): aggressive behavior [26] and internalizing problems [27] (downloaded on February 10th 2017); (e) Anxiety Neuro Genetics Study: anxiety disorder [28] (downloaded on November 28th 2016), (f) International Genomics of Alzheimer's Project (IGAP): Alzheimer's disease [29] (downloaded on February 12th 2017).
In case of unavailability of the identified metabolite SNPs in the respective 14 GWAS and GWAMA, we used proxy-SNPs. The SNPs with minimum linkage disequilibrium (LD) r2 ≥ 0.80 on the basis of 1000 Genomes, Phase 3 (Oct. 2014) for European ancestry (maximal distance of 500 kb) were exported applying the in silico tool rAggr (University of Southern California (USC; http://raggr.usc.edu/). Post-hoc selection criteria for proxy-SNPs were defined: 1st highest r2, 2nd smallest distance to the lead SNP. To define separate chromosomal regions we used a distance criterion of +/−500 kb surrounding the middle SNP [7] of a chromosomal locus. Consecutive numbers were assigned to each region.
Independent data for the confirmation of SNPs identified to be shared between metabolic and mental traits were available for (a) BMI (GWAS data of Japanese individuals (n = 173,430; [6]; data downloaded on September 25th 2017 from https://humandbs.biosciencedbc.jp/en/hum0014-v6#JGAS00000000114%20/%20hum 0014.v6.158k.v1) and (b) Educational Attainment (UK Biobank sample (n = 111,114; [30]; data downloaded on December 4th 2017 from https://grasp.nhlbi.nih.gov/FullResults.aspx).
2.2. Statistical analyses
To control for the overall type I error rate in the lookup, a Bonferroni correction was used. We performed a conservative study-wide correction for 516 SNPs and all 14 traits which yielded a threshold p-value of 6.92 × 10−6. We additionally assumed a univariate multiple regression model for each psychiatric trait entailing a Bonferroni correction for 516 tests (=516 SNPs), resulting in a less strict threshold p-value of 9.7 × 10−5. For both corrections, independence of SNPs was assumed. However, although the selected dataset only included 216 chromosomal regions, we did not prune SNPs in high LD to adhere to the most conservative approach possible.
Additionally, we compared the number of observed associations between metabolite SNPs or their proxies and mental phenotypes with the expected number as based on the null hypothesis (no association; see Table 2). For this analysis, we assumed that the 516 SNPs were located in 216 chromosomal regions (1 Mb each). For each mental phenotype, the expected total number of false positive hits was computed as follows: 0.001 × 216 = 0.216 for a p-value ≤ 0.001. For confirmatory analyses for BMI and educational attainment a p-value < 0.05 was considered as significant.
Table 2.
Comparison between observed and expected hits stratified by phenotype.
| Nominal p-value | Expected hits per phenotype A | Number of hits per phenotype |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI | Ed_Att | Neuro-ticism | Alzheimer | Schizo-phrenia | Well-being | Aggressive behavior | Anxiety | Depressive Symptoms | MDD | Bipolar disorder | Autism | Internalizing problems | ADHD | ||
| p ≤ 0.001 | 0.216 | 11 | 8 | 2 | 2 | 14 | 1 | 1 | 4 | 2 | |||||
| p ≤ 0.0001 | 0.0216 | 9 | 4 | 2 | 9 | 1 | 1 | ||||||||
| p ≤ 0.00001 | 0.00216 | 5 | 1 | 1 | 3 | 1 | |||||||||
| p ≤ 0.000001 | 0.000216 | 2 | 1 | 1 | |||||||||||
| p ≤ 0.0000001 | 0.0000216 | 2 | 1 | ||||||||||||
| p ≤ 0.00000001 | 0.00000216 | 1 | 1 | ||||||||||||
Expected number of false positive hits was computed as follows: 0.001 x 216=0.216 for p-value≤0.001.
ADHD Attention Deficit/Hyperactivity Disorder; BMI, Body Mass Index; Ed_Att: Educational attainment.
Based on the identified 216 chromosomal regions of 1 MB (see Methods online).
3. Results
3.1. Observed associations linking human metabolism with BMI and mental traits/disorders
Our search identified 516 SNPs that were genome-wide significantly associated with metabolite levels and ratios thereof in human blood and/or urine (Figure 1) in GWAS or GWAMA of human metabolism (Supplemental Table 1). Of these, 19 SNPs were significantly associated with specific mental phenotypes when using the strict Bonferroni correction for both 516 SNPs and 14 traits (p < 6.92 × 10−6; Table 1) in GWAS and GWAMA of BMI and mental traits/disorders (Supplemental Table 2). The less conservative adjustment for the 516 SNPs merely within each phenotype resulted in a total of 41 SNPs that met the Bonferroni-corrected threshold (p < 9.69 × 10−5). The numbers of identified hits per phenotype (p < 6.92 × 10−6) correlated with the known numbers of genome-wide significant findings (considered as percentages of examined SNPs) for the respective datasets (Spearman rank correlation: r = 0.72 p = 0.003).
Table 1.
Associations of GWAS and GWAMA derived human metabolism SNPs in GWAMA data for mental disorders and other mental/behavioral phenotypes.
| SNP | Region | Mapped Gene | Effect allele | MetaboliteA | p-value for metabolite or metabolite ratio | Exponents of p-values per mental phenotype |
Effect directionsB | Source | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI | Ed_Att | Neuroticism | Alzheimer | Schizophrenia | Well-being | Aggression | Anxiety | Depression | MDD | Bipolar | Autism | Internalizing | ADHD | ||||||||
| rs2802729 | 1q43 | SDCCAG8 | A | eGFRcrea | 2.0E-8 | −3 | −6* | −2 | −3 | −2 | A: metab↓; Schizo↓ | [31] | |||||||||
| rs6546838 | 2p13.1 | ALMS1 | A | eGFRcrea | 8.0E-20 | −3 | −6* | A: metab ↓; Schizo↓ | [31] | ||||||||||||
| rs9309473 | 2p13.1 | ALMS1 | G | N-acetylated compound(s) | 4.1E-19 | −3 | −6* | G: metab↑; Schizo↑ | [46] | ||||||||||||
| rs11884776 | 2p13.1 | ALMS1 | C | N-acetylated compounds | 6.0E-17 | −3 | −6* | C: metab↑; Schizo↓ | [45] | ||||||||||||
| rs10469966 | 2p13.1 | ALMS1 | A | X-12093 | 1.0E-51 | −4 | −7* | A: metab↑; Schizo↑ | [13] | ||||||||||||
| rs6546847 | 2p13.1 | ALMS1 | A | N-acetylated compounds | 5.3E-161 | −3 | −6* | A: metab↑; Schizo↓ | [45] | ||||||||||||
| rs13391552 | 2p13.1 | ALMS1 | A | N-acetylornithine | 5.0E-252 | −3 | −6* | A: metab↓; Schizo↑ | [63] | ||||||||||||
| rs6546857 | 2p13.1 | ALMS1 - NAT8 | A | (X-11787) | 1.0E-23 | −3 | −7* | A: metab↑; Schizo↓ | [64] | ||||||||||||
| rs10178409 | 2p13.1 | ALMS1 - NAT8 | T | N-acetylaspartate | 1.0E-95 | −3 | −5 | T: metab↑; Schizo↑ | [65] | ||||||||||||
| rs13538 | 2p13.1 | NAT8 | A | N-acetylornithine/myo-inositol | 8.0E-157 | −3 | −5 | A: metab↑; Schizo↓ | [13] | ||||||||||||
| rs13538 | 2p13.1 | NAT8 | G | eGFRcrea | 5.0E-14 | −3 | −5 | G: metab↑; Schizo↑ | [66] | ||||||||||||
| rs13538 | 2p13.1 | NAT8 | A | X-11787 | 2.0E-23 | −3 | −5 | A: metab↑; Schizo↓ | [64] | ||||||||||||
| rs10206899 | 2p13.1 | ALMS1P | T | N-acetylornithine | 2.0E-14 | −3 | −5 | T: metab↑; Schizo↓ | [13] | ||||||||||||
| rs1260326 | 2p23.3 | GCKR | A | HDL total | 6.3E-36 | −5 | −3 | −3 | A: metab↑;BMI↓ | [67] | |||||||||||
| rs1260326 | 2p23.3 | GCKR | n.a. | S-HDL-P | 1.0E-12 | −5 | −3 | −3 | ? | [68] | |||||||||||
| rs1260326 | 2p23.3 | GCKR | C | mannose | 6.0E-56 | −5 | −3 | −3 | C: metab↑; BMI↑ | [69] | |||||||||||
| rs1260326 | 2p23.3 | GCKR | T | eGFRcrea | 3.0E-14 | −5 | −3 | −3 | T: metab↑; BMI↓ | [66] | |||||||||||
| rs1260326 | 2p23.3 | GCKR | T | glucose/mannose | 3.0E-148 | −5 | −3 | −3 | T: metab↑; BMI↓ | [13] | |||||||||||
| rs7570971 | 2q21.3 | RAB3GAP1 | A | 1.5-anhydroglucitol | 8.0E-45 | −6* | −2 | A: metab↓; BMI ↓ | [13] | ||||||||||||
| rs1047891 | 2q34 | CPS1 | A | plasma homocysteine levels | 9.0E-13 | −5 | A: metab↑; BMI↑ | [70] | |||||||||||||
| rs715 | 2q34 | CPS1 | T | serine | 3.0E-11 | −6 | T: metab↓; BMI↓ | [71] | |||||||||||||
| rs715 | 2q34 | CPS1 | T | glycine | 3.0E-50 | −6 | T: metab↓; BMI↓ | [13],[72],[65] | |||||||||||||
| rs10513801 | 3q27.2 | ETV5 | T | eGFRcrea | 1.0E-9 | −21* | −5 | T: metab↑; BMI↑; EdAtt↑ | [31] | ||||||||||||
| rs12654264 | 5q13.3 | HMGCR | T | LDLc | 1.0E-20 | −8* | −3 | −2 | −2 | T: metab ↓; BMI↓ | [58] | ||||||||||
| rs7759001 | 6p22.1 | ZNF204P | A | eGFRcrea | 2.0E-8 | −2 | −5 | −2 | −3 | A: metab↓; Schizo↓ | [31] | ||||||||||
| rs2762353 | 6p22.2 | SLC17A1 | A | 4-androsten-3beta,17beta-diol disulfate 2 | 3.0E-13 | −5 | −3 | A; metab↑; Schizo↑ | [13] | ||||||||||||
| rs9400467 | 6q21 | SLC16A10 | T | tyrosine | 7.0E-14 | −6* | T: metab ↓; Schizo↓ | [13],[71] | |||||||||||||
| rs4841132 | 8p23.1 | LOC157273 | n.a. | free cholesterol in medium HDL | 2.0E-9 | −3 | −5 | n.a. | [68] | ||||||||||||
| rs15676 | 9q34.11 | TBC1D13 | A | indolelactate | 1.0E-12 | −3 | −5 | −2 | A: metab↑; Schizo↑ | [13] | |||||||||||
| rs9527 | 10q24.32 | C10orf32 C10orf32-ASMT | A | dimethylarsinic acid in urine | 3.0E-9 | −2 | −5 | n.a. | [43] | ||||||||||||
| rs1278587 | 11q13.4 | NADSYN1 | T | vitamin D insufficiency | 2.0E-27 | −5 | n.a. | [73] | |||||||||||||
| rs3184504 | 12q24.12 | SH2B3 | T | kynurenine | 6.0E-18 | −6 | T: metab↑; BMI↓ | [13] | |||||||||||||
| rs2066938 | 12q24.31 | UNC119B | A | butyrylcarnitine | 3.1E-630 | −6* | −4 | A: metab↓; EdAtt↑ | [13] | ||||||||||||
| rs2066938 | 12q24.31 | UNC119B | A | butyrylcarnitine /propionylcarnitine | 4.4E-305 | −6* | −4 | A: metab↓; EdAtt↑ | [63] | ||||||||||||
| rs2014355 | 12q24.31 | ACADS | T | C3/C4 | 5.0E-96 | −6* | −3 | T: metab↓; edAtt↑ | [12] | ||||||||||||
| rs3916 | 12q24.31 | ACADS | G | unknown | 2.4E-22 | −6* | −3 | n.a. | [45] | ||||||||||||
| rs4144027 | 14q32.33 | LOC105370690 | T | aspargine | 1.0E-11 | −2 | −5 | −2 | −2 | −2 | T: metab↑; Schizo↓ | [13] | |||||||||
| rs12446492 | 16p12.3 | PDILT | A | uromodulin indexed to creatinine | 6.0E-27 | −5 | A: metab↓; BMI↑ | [74] | |||||||||||||
| rs7200543 | 16p13.11 | PDXDC1 | A | PC aa C38:3 (Glycerophospholipid levels) | 3.0E-17 | −5 | −3 | −2 | A: metab↑; BMI↑ | [71] | |||||||||||
| rs7200543 | 16p13.11 | PDXDC1 | G | 1-eicosatrienoylglycero-phosphocholine /1-linoleoylglycero-phosphocholine | 5.0E-16 | G: metab↓; BMI↓ | [63] | ||||||||||||||
| rs11075253 | 16p13.11 | PDXDC1, NTAN1 | A | linoleic acid/PUFA | 5.0E-15 | −5 | −2 | −2 | −2 | A: metab↓; EdAtt↑ | [75] | ||||||||||
| rs8056893 | 16q22.1 | SLC7A6 | A | glutaroyl carnitine | 2.0E-30 | −5 | −2 | −2 | A: metab↓; BMI↓ | [13] | |||||||||||
| rs2863979 | 16q22.1 | SLC7A6 | A | lysine | 1.0E-17 | −5 | −2 | −2 | −2 | A: metab↑; BMI↓ | [13] | ||||||||||
| rs9916302 | 17q12 | FBXL20 | T | eGFRcrea | 5.0E-15 | −5 | −3 | T: metab↓; EdAtt↑ | [31] | ||||||||||||
| rs7219014 | 17q12 | CDK12 | A | histidine/τ-methylhistidine | 4.0E-26 | −5 | −3 | A: metab↓; EdAtt↓ | [65] | ||||||||||||
| rs4808136 | 19p13.11 | ELL | A | myo-inositol | 5.0E-14 | −3 | −5 | −2 | −3 | −2 | A: metab↑; Alz↑ | [13] | |||||||||
| rs4803750 | 19q13.32 | BCL3 | G | LDL-C assay fasting | 1.0E-27 | −13* | G: metab↓; Alz↓ | [67] | |||||||||||||
| rs7412 | 19q13.32 | APOE | T | L-LDL-FC | 3.0E-58 | −2 | −22* | T: metab↓; Alz ↓ | [75] | ||||||||||||
| rs4420638 | 19q13.32 | APOC1- APOC1P1 | A | LDL-C | 1.0E-14 | −4 | −454* | −2 | G: metab↑; Alz↑ proxy | [76] | |||||||||||
| rs2287921 | 19q13.33 | RASIP1 | C | FUT2 - fucose | 7.0E-19 | −2 | −2 | −2 | −3 | −6* | C: metab↑, BIP↑ | [45] | |||||||||
Horizontal lines separate chromosomal regions of 1 Mb.
Bold values indicate significant results upon correction for 516 SNPs (p < 9.7 × 10-5).
*Significant values upon correction for 14 traits and 516 SNPs (p < 6.92 × 10-6).
ADHD: Attention Deficit/Hyperactivity Disorder; Aggression: Aggressive behavior; Alz: Alzheimer's disease; Autism: Autism spectrum disorder; Bipolar: Bipolar disorder; BMI: Body mass index; Ed_Att: Educational attainment; Internalizing: Internalizing problems; GWAS: Genome-wide association study; GWAMA: Genome-wide association meta-analysis; MDD: Major depressive disorder, metab: Metabolite or ratio of metabolites.
Metabolite, ratio of metabolites, or metabolism markers with lowest p-value in GWAMA.
Upward arrows indicate positive beta-values or odds ratios >1, downward arrows negative beta-values or odds ratios <1 for the phenotype showing the lowest p-value.
Based on our definition of independent loci (see Methods), the 516 SNPs belong to 216 regions. Accordingly, the 19 and 41 SNPs are located in nine and 24 regions, respectively (Table 1, Supplemental Table 4). Hence, approximately 5% (10% upon use of the less conservative threshold of p < 9.69 × 10−5) of the 216 regions which are relevant for the regulation of blood/urine metabolite levels also appear to play a role in mental disorders and related quantitative phenotypes. The largest numbers of hits were observed for BMI, schizophrenia, and educational attainment (Table 1, Table 2).
Genetic variation associated with estimated glomerular filtration rate (eGFR), a marker for renal function, which is predominantly calculated based on serum creatinine levels, appeared frequently in our hits for schizophrenia (three loci with p < 9.69 × 10−5), educational attainment (two loci) and BMI (two loci). Of the 53 loci detected in the most recent GWAMA for eGFR [31] 43 loci (81%) were nominally associated (p < 0.05) with one or more of the investigated phenotypes (Supplemental Table 4).
3.2. Cross–phenotype associations
Among the 41 SNPs located in the 24 regions, ETV5 was the only locus that met our less strict threshold of p < 9.7 × 10−5 for more than one phenotype, i.e. BMI and educational attainment. However, the potential cross-phenotype relevance of specific loci is illustrated by the fact that for four, five (twice), and 20 of the 41 SNPs, nominal hits (p < 0.05) were found for a total of four, three, two and one additional phenotypes, respectively (Table 1). Interestingly, four of the nine regions identified for schizophrenia (p < 9.69 × 10−5) were nominally also associated with educational attainment (vice versa two of four). For the phenotypes MDD, depressive symptoms, anxiety, ASD, well-being, and neuroticism, we observed no genetic cross-link with human metabolism.
3.3. Confirmatory analyses
The confirmatory analyses (Table 3) in independent samples comprised all SNPs that fulfilled the p-value threshold of p < 9.69 × 10−5 for BMI or educational attainment in the discovery analyses, respectively. For BMI five of the nine regions with p < 9.69 × 10−5 in the discovery data set revealed p-values < 0.05 in the confirmatory data set based on Japanese subjects. For educational attainment, three of four regions in the discovery data set revealed p-values < 0.05 in the confirmatory data set (UK Biobank). The directions of all effect alleles with were consistent between discovery and confirmatory data sets.
Table 3.
Confirmation of SNPs for BMI and educational attainment fulfilling the p-value treshold p < 9.69 × 10−5.
| SNP | Region | Mapped Gene | Effect allele | P-values |
|||
|---|---|---|---|---|---|---|---|
| BMI – Discovery | BMI - Confirmation | Educational Attainment - Discovery | Educational Attainment - Confirmation | ||||
| rs1260326 | 2p23.3 | GCKR | A | 9.2 × 10−5 | n.s. | ||
| rs7570971 | 2q21.3 | RAB3GAP1 | A | 1.5 × 10−6 | n.s. | ||
| rs1047891 | 2q34 | CPS1 | A | 8.1 × 10−5 | 4.7 × 10−3 | ||
| rs715 | 2q34 | CPS1 | T | 7.1 × 10−6 | 4.7 × 10−3 | ||
| rs10513801 | 3q27.2 | ETV5 | T | 1.1 × 10−21 | 3.4 × 10−3 | 2.0 × 10−5 | 1.3 × 10−2 |
| rs12654264 | 5q13.3 | HMGCR | T | 1.8 × 10−8 | 8.3 × 10−9 | ||
| rs3184504 | 12q24.12 | SH2B3 | T | 9.3 × 10−6 | n.s. | ||
| rs2066938 | 12q24.31 | UNC119B | A | 4.4 × 10−6 | n.s. | ||
| rs2014355 | 12q24.31 | ACADS | T | 5.0 × 10−6 | n.s. | ||
| rs3916 | 12q24.31 | ACADS | G | 3.2 × 10−6 | n.s. | ||
| rs12446492 | 16p12.3 | PDILT | A | 3.0 × 10−5 | n.s. | ||
| rs7200543 | 16p13.11 | PDXDC1 | A | 1.0 × 10−5 | 4.2 × 10−5 | ||
| rs11075253 | 16p13.11 | PDXDC1, NTAN1 | A | 4.6 × 10−5 | 7.0 × 10−4 | ||
| rs8056893 | 16q22.1 | SLC7A6 | A | 3.1 × 10−5 | 4.5 × 10−3 | ||
| rs2863979 | 16q22.1 | SLC7A6 | A | 2.7 × 10−5 | 3.6 × 10−3 | ||
| rs9916302 | 17q12 | FBXL20 | T | 1.9 × 10−5 | 3.9 × 10−2 | ||
| rs7219014 | 17q12 | CDK12 | A | 1.1 × 10−5 | 1.6 × 10−2 | ||
Horizontal lines separate chromosomal regions of 1 Mb.
Bold values indicate significant values in the discovery data sets upon correction for 14 traits and 516 SNPs (p < 6.92 × 10−6).
BMI: Body mass index; n.s: p ≥ 0.05.
The effect allele direction was consistent in discovery and confirmatory data for all replicated SNPs.
4. Discussion
The main finding of the current study was the identification of SNPs that both explain variation of human metabolite concentrations and are associated with BMI and mental traits/disorders. Approximately 5–10% of the regions, which are relevant for the regulation of blood/urine metabolite levels, were also associated with BMI/mental phenotypes. Furthermore, our approach re-identified three genome-wide significant loci (p ≤ 5 × 10−8) in the respective data sets, i.e. ETV5 [32] and HMGCR [7] for BMI, and APO-E4 for Alzheimer's disease [29]). Two further loci (C10orf32-AS3MT and SDCCAG8) surpassed the genome-wide significance level in analyses for schizophrenia [21], [33]. However, in the available GWAMA dataset (see Methods), the respective SNPs in these two regions had not reached genome-wide significance.
For schizophrenia and educational attainment, our examination revealed several loci with cross-phenotype relevance. Positive but low genetic correlations (rg = 0.09–0.10, p < 0.05) between schizophrenia and educational attainment have been reported previously [1], [11], [34]. Overall, the genetic link between metabolism and mental phenotypes was mainly apparent for BMI, schizophrenia, and educational attainment. The number of significant hits for these three phenotypes seemingly parallels the high number of previously identified genome-wide significant findings (Supplemental Table 1). Conversely, in the GWAS and GWAMA datasets Aggressive Behavior, Anxiety, Depressive Symptoms, MDD, ASD, and Internalizing Problems, nominal p-values ≤ 0.001 were not detected (Table 2). For these phenotypes genome-wide significant findings had not been reported in the analyzed data sets. The lack of overlap between SNPs involved in metabolite regulation with aggressive behavior, anxiety, depressive symptoms, MDD, ASD, and internalizing problems might indicate that genetic variations in human metabolism are of minor relevance for these phenotypes, not disputing that metabolic disturbances – albeit not genetically dominated - may well impact the respective mental trait. However, the lack of significant hits might also reflect insufficient statistical power due to the relatively small sample sizes for MDD, anxiety, and ASD in particular. For future analyses of a similar type, we recommend focusing on datasets with at least one genome-wide significant finding. Unfortunately, the data set of the most recent GWAMA for MDD [35] was not available for download on the PGC website at the time of the lookup; it will be of particular interest to analyze this data set to determine if metabolite SNPs overlap between BMI and MDD.
One important finding was that genetic variation associated with eGFR seems to play a relevant role in our hits for schizophrenia. Reduced renal clearance of specific toxins had been postulated to elevate the risk for schizophrenia decades ago; however, small-scaled efforts to treat schizophrenia with dialysis failed [36]. More recently, schizophrenia has been shown to be associated with an increased risk of chronic kidney disease (CKD) [37]. However, for all three of the identified eGFR loci relevant for schizophrenia the observed effect directions of the respective SNPs were not consistent with the hypothesis that a reduced eGFR might causally underlie these findings. LSCD analyses (http://ldsc.broadinstitute.org/) also revealed a negative genetic correlation (rg = −0.166, p = 0.016) between CKD and schizophrenia, but a slightly positive correlation between schizophrenia and serum creatinine (rg = 0.067, p = 0.028). Similarly, for educational attainment, the direction of effect of the two eGFR SNPs again appears counterintuitive since reduced renal function is clinically associated with cognitive deficits: cognitive deficits develop early in CKD and worsen over time independently of other risk factors [38]. Clinically, the encephalopathy observed in end-stage renal disease is probably caused by the accumulation of uremic toxins [39]. Overall, our results do not support the notion from clinical observations that metabolite SNPs that reduce eGFR are associated with an increased risk of schizophrenia or a lower level of educational attainment.
Toxins from e.g. environmental pollution (e.g. so-called endocrine disruptors like phthalates and chlorinated polyphenols or heavy metal exposures) are potential contributors to the etiology of obesity [40], [41] and schizophrenia [42]. Our analysis did not support this hypothesis for obesity; for schizophrenia, we identified genetic variation in the C10orf32-AS3MT gene region (Supplemental Table 3) as one potential cross-link between arsenic detoxification metabolism and schizophrenia; the respective locus was identified in a GWAS of arsenic metabolism and toxicity phenotypes in Bangladesh [43]. Furthermore, the gene product of N-acetyltransferase 8 (NAT8), which is located in the same chromosomal region, is involved in xenobiotic metabolism and detoxification processes [44]. However, SNPs near NAT8, which increase levels of N-acetylated compounds, appear to have opposite effects on schizophrenia (e.g. rs9309473 and rs11884776; Table 1), which potentially reflects inconsistently reported effect directions in the underlying studies [21], [45], [46], or point speculatively towards independent roles in brain versus somatic pathways.
The identification of biomarkers [47] may improve diagnostic classification systems of mental disorders and related phenotypes [48]. The current lack of biomarkers represents a key challenge for psychiatric research according to the U.S. National Institute of Mental Health's (NIMH) new Research Domain Criteria (RDoC) [49]. This study, for the first time, analyzed the overlap between SNPs involved in the regulation of blood/urine metabolites and mental phenotypes at the DNA level on a large scale including 14 phenotypes. Therefore, our approach could represent a step towards the identification of genetically based biomarkers, thus assisting in the classification of biologically more homogenous subtypes of mental disorders [50].
Based on our results, the evaluation of the identified serum/urine metabolites in appropriately characterized and sufficiently powered (patient) samples represents an initial step towards investigating a potential causal link between the respective metabolite concentrations and the corresponding phenotype(s). In fact, for some structural classes of steroids, which have been identified in the current study, differences in steroid levels between patients with schizophrenia and controls have already been observed [51].
However, a note of caution is warranted. The identified SNPs may have a link with the respective phenotype via mechanisms that are not related to the serum/urine concentrations of the respective metabolite. For example, an enzyme could be differentially regulated in the periphery and the central nervous system, with only the latter explaining the genetic predisposition to the mental phenotype. Alternatively, some of the findings may prove to be due to a disease locus being in LD with the respective metabolite locus. Therefore, case–control studies of the metabolite concentrations tagged by the SNPs listed in Table 1 are needed to follow-up on our findings, e.g. on markers for kidney function (e.g. eGFR) and schizophrenia, BMI, and educational attainment. However, the strength of our approach is that the genetic impact observed for the identified metabolites indicates that at least part of the variance in metabolite levels does not seem to result from the mental phenotype itself or from medications but could parallel or precede the onset of the mental phenotype.
To evaluate any causal or pleiotropic effects of the metabolites on BMI/mental phenotypes identified in our lookup, we suggest subsequent studies using two-sample Mendelian Randomization [52], [53], [54], [55] with multiple SNPs or a polygenic risk score with a strong association to a metabolite as instrumental variables. A specific challenge here would be that Mendelian Randomization assumes no pleiotropy, but many of the SNPs identified in our study are associated with a number of metabolites of interest. To address this issue, the recently suggested multivariable Mendelian Randomization approach could be employed that was shown to be successful in disentangling different lipid pathways in their causal roles for coronary heart disease [56].
A further limitation of our work pertains to the identification of metabolites of interest via the GWAS Catalog. Single GWAS reports for metabolite concentrations were not covered by this website [57]. Furthermore, these GWAS differed with regard to ancestry of the study population, e.g. Kim et al. focused on East-Asians population [58]. Therefore, genetic ancestry could be a potential confounding factor. Another limitation is that for our confirmatory analyses we found independent datasets for only BMI and educational attainment. Because the BMI data sets are based on subjects of different ethnic origin, the confirmation of BMI loci in the Japanese data set suggests a common predisposition in both Europeans and the Japanese. Further replication analyses are necessary to confirm the role of the identified metabolites in light of p-values that did not meet the criterion of genome-wide significance established for GWAS or GWAMA. However, the detection of the five previously identified loci with p-values < 5 × 10−8 seemingly indirectly “validates” our approach.
5. Conclusion
There is a substantial genetic overlap between metabolism, BMI, and mental phenotypes, as 5–10% of the regions, which are relevant for the regulation of blood/urine metabolite levels, were significantly associated with BMI and mental phenotypes.
Considering results from this study and previous clinical observations [38], [39], [59], [60], [61], [62], the feasibility of eGFR, acetylated-compounds, and heavy-metal metabolites as potential biomarkers needs to be examined in case–control studies, especially in schizophrenia. A diagnostic use of these potential biomarkers and an adjustment of metabolite levels e.g. via dietary interventions could guide prevention and additional intervention concepts for obesity and schizophrenia. Assuming a confirmation of these findings in case–control studies, metabolic traits related to these biomarkers might even help the development of future treatment options. If some of the tagged genes are involved in detoxification, the disease risk alleles may prove to be relevant under specific environmental conditions only. In such scenarios, the effect sizes may prove to be more substantial than the current findings suggest, which could become relevant for efficacious targeted therapeutic and preventive efforts.
Acknowledgements
The authors express their gratitude to all participants. Additionally we like to thank the different consortia for providing publically available GWAMA data sets: (a) the Psychiatric Genetics Consortium (PGC), (b) the Social Science Genetic Association Consortium (SSGAC), (c) the Genetic Investigation of Anthropometric Traits (GIANT) Consortium, (d) Early Genetics and Lifecourse Epidemiology Consortium (EAGLE), (e) Anxiety Neuro Genetics Study, (f) International Genomics of Alzheimer's Project (IGAP), (g) BMI GWAS data of Japanese individuals and (h) UK Biobank sample. We thank the following sources for funding or research: the German Ministry for Education and Research (National Genome Research Net-Plus 01GS0820), the German Research Foundation (DFG; HI865/2-1), the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreements n°245009 and n°262055. Part of this work was initiated in ECNP Nutrition Network.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.03.015.
Conflict of interest
Dr. Correll has been a consultant and/or advisor to or has received honoraria from: Alkermes, Allergan, Bristol-Myers Squibb, Gerson Lehrman Group, IntraCellular Therapies, Janssen/J&J, LB Pharma, Lundbeck, Medavante, Medscape, Neurocrine, Otsuka, Pfizer, ROVIA, Sunovion, Takeda, and Teva. He has provided expert testimony for Bristol-Myers Squibb, Janssen, and Otsuka. He served on a Data Safety Monitoring Board for Lundbeck, Pfizer and ROVIA. He received royalties from UpToDate and grant support from Janssen, Neurocrine and Takeda.
All other authors report no biomedical financial interests or potential conflicts of interest.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R. An atlas of genetic correlations across human diseases and traits. Nature Genetics. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okbay A., Baselmans B.M., De Neve J.E., Turley P., Nivard M.G., Fontana M.A. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature Genetics. 2016;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okbay A., Baselmans B.M., De Neve J.E., Turley P., Nivard M.G., Fontana M.A. Corrigendum: genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature Genetics. 2016;48:1591. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okbay A., Baselmans B.M., Neve J.E., Turley P., Nivard M.G., Fontana M.A. Corrigendum: genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature Genetics. 2016;48:970. doi: 10.1038/ng0816-970c. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama M., Okada Y., Kanai M., Takahashi A., Momozawa Y., Ikeda M. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nature Genetics. 2017;49:1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 7.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avila C., Holloway A.C., Hahn M.K., Morrison K.M., Restivo M., Anglin R. An overview of links between obesity and mental health. Current Obesity Reports. 2015;4:303–310. doi: 10.1007/s13679-015-0164-9. [DOI] [PubMed] [Google Scholar]
- 9.Hinney A., Kesselmeier M., Jall S., Volckmar A.L., Focker M., Antel J. Evidence for three genetic loci involved in both anorexia nervosa risk and variation of body mass index. Molecular Psychiatry. 2017;22:321–322. doi: 10.1038/mp.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho J.E., Larson M.G., Ghorbani A., Cheng S., Chen M.H., Keyes M. Metabolomic profiles of body mass index in the framingham heart study reveal distinct cardiometabolic phenotypes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148361. e0148361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anttila V., Bulik-Sullivan B., Finucane H., Bras J., Duncan L., Escott-Price V. Analysis of shared heritability in common disorders of the brain. Preprint at bioRxiv. 2016 doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., Prehn C. A genome-wide perspective of genetic variation in human metabolism. Nature Genetics. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin S.Y., Fauman E.B., Petersen A.K., Krumsiek J., Santos R., Huang J. An atlas of genetic influences on human blood metabolites. Nature Genetics. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozman J., Rathkolb B., Oestereicher M.A., Schutt C., Ravindranath A.C., Leuchtenberger S. Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nature Communications. 2018;9:288. doi: 10.1038/s41467-017-01995-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halama A., Horsch M., Kastenmuller G., Moller G., Kumar P., Prehn C. Metabolic switch during adipogenesis: from branched chain amino acid catabolism to lipid synthesis. Archives of Biochemistry and Biophysics. 2016;589:93–107. doi: 10.1016/j.abb.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Kageyama Y., Kasahara T., Morishita H., Mataga N., Deguchi Y., Tani M. Search for plasma biomarkers in drug-free patients with bipolar disorder and schizophrenia using metabolome analysis. Psychiatry and Clinical Neurosciences. 2017;71:115–123. doi: 10.1111/pcn.12461. [DOI] [PubMed] [Google Scholar]
- 17.Koike S., Bundo M., Iwamoto K., Suga M., Kuwabara H., Ohashi Y. A snapshot of plasma metabolites in first-episode schizophrenia: a capillary electrophoresis time-of-flight mass spectrometry study. Translational Psychiatry. 2014;4:e379. doi: 10.1038/tp.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt A., Martins-de-Souza D., Akbarian S., Cassoli J.S., Ehrenreich H., Fischer A. Consensus paper of the WFSBP Task Force on Biological Markers: criteria for biomarkers and endophenotypes of schizophrenia, part III: molecular mechanisms. The World Journal of Biological Psychiatry. 2017;18:330–356. doi: 10.1080/15622975.2016.1224929. [DOI] [PubMed] [Google Scholar]
- 19.Brookes K.J., Chen W., Xu X., Taylor E., Asherson P. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biological Psychiatry. 2006;60:1053–1061. doi: 10.1016/j.biopsych.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 20.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripke S. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Major Depressive Disorder Working Group of the Psychiatric G. C, Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neale B.M., Medland S.E., Ripke S., Asherson P., Franke B., Lesch K.P. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sklar P. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium . March 2015. Dataset: PGC-ASD summary statistics from a meta-analysis of 5,305 ASD-diagnosed cases and 5,305 pseudocontrols of European descent (based on similarity to CEPH reference genotypes)http://www.med.unc.edu/pgc/results-and-downloads available at: [Google Scholar]
- 26.Pappa I., St Pourcain B., Benke K., Cavadino A., Hakulinen C., Nivard M.G. A genome-wide approach to children's aggressive behavior: The EAGLE consortium. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2016;171:562–572. doi: 10.1002/ajmg.b.32333. [DOI] [PubMed] [Google Scholar]
- 27.Benke K.S., Nivard M.G., Velders F.P., Walters R.K., Pappa I., Scheet P.A. A genome-wide association meta-analysis of preschool internalizing problems. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53:667–676. doi: 10.1016/j.jaac.2013.12.028. e7. [DOI] [PubMed] [Google Scholar]
- 28.Otowa T., Hek K., Lee M., Byrne E.M., Mirza S.S., Nivard M.G. Meta-analysis of genome-wide association studies of anxiety disorders. Molecular Psychiatry. 2016;21:1391–1399. doi: 10.1038/mp.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature Genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies G., Marioni R.E., Liewald D.C., Hill W.D., Hagenaars S.P., Harris S.E. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Molecular Psychiatry. 2016;21:758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattaro C., Teumer A., Gorski M., Chu A.Y., Li M., Mijatovic V. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nature Communications. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berndt S.I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M.F. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nature Genetics. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warrier V., Bethlehem R.A.I., Geschwind D., Baron-Cohen S. Genetic overlap between educational attainment, schizophrenia and autism. Preprint at bioRxiv. 2016 https://www.biorxiv.org/content/early/2016/12/12/093575 [Google Scholar]
- 35.Wray N., Sullivan P. Major depressive disorder working Group of the PGC. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Preprint at bioRxiv. 2017 doi: 10.1038/s41588-018-0090-3. https://www.biorxiv.org/content/early/2017/07/24/167577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroll P., Port F.K., Silk K.R. Hemodialysis and schizophrenia. A negative report. The Journal of Nervous and Mental Disease. 1978;166:291–293. doi: 10.1097/00005053-197804000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Tzeng N.S., Hsu Y.H., Ho S.Y., Kuo Y.C., Lee H.C., Yin Y.J. Is schizophrenia associated with an increased risk of chronic kidney disease? A nationwide matched-cohort study. BMJ open. 2015;5 doi: 10.1136/bmjopen-2014-006777. e006777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai S., Mecarelli O., Pulitano P., Romanello R., Davi L., Zarabla A. Neurological, psychological, and cognitive disorders in patients with chronic kidney disease on conservative and replacement therapy. Medicine. 2016;95 doi: 10.1097/MD.0000000000005191. e5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seifter J.L., Samuels M.A. Uremic encephalopathy and other brain disorders associated with renal failure. Seminars in Neurology. 2011;31:139–143. doi: 10.1055/s-0031-1277984. [DOI] [PubMed] [Google Scholar]
- 40.Hyman M. Systems biology, toxins, obesity, and functional medicine. Alternative Therapies in Health and Medicine. 2007;13:S134–S139. [PubMed] [Google Scholar]
- 41.Elobeid M.A., Allison D.B. Putative environmental-endocrine disruptors and obesity: a review. Current Opinion in Endocrinology Diabetes and Obesity. 2008;15:403–408. doi: 10.1097/MED.0b013e32830ce95c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Attademo L., Bernardini F., Garinella R., Compton M.T. Environmental pollution and risk of psychotic disorders: a review of the science to date. Schizophrenia Research. 2017;181:55–59. doi: 10.1016/j.schres.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Pierce B.L., Kibriya M.G., Tong L., Jasmine F., Argos M., Roy S. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002522. e1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veiga-da-Cunha M., Tyteca D., Stroobant V., Courtoy P.J., Opperdoes F.R., Van Schaftingen E. Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids. Journal of Biological Chemistry. 2010;285:18888–18898. doi: 10.1074/jbc.M110.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rueedi R., Ledda M., Nicholls A.W., Salek R.M., Marques-Vidal P., Morya E. Genome-wide association study of metabolic traits reveals novel gene-metabolite-disease links. PLoS Genetics. 2014;10 doi: 10.1371/journal.pgen.1004132. e1004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholson G., Rantalainen M., Li J.V., Maher A.D., Malmodin D., Ahmadi K.R. A genome-wide metabolic QTL analysis in Europeans implicates two loci shaped by recent positive selection. PLoS Genetics. 2011;7 doi: 10.1371/journal.pgen.1002270. e1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biomarkers Definitions Working G Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical Pharmacology and Therapeutics. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 48.Niciu M.J., Mathews D.C., Ionescu D.F., Richards E.M., Furey M.L., Yuan P. Biomarkers in mood disorders research: developing new and improved therapeutics. Revista de psiquiatria clinica. 2014;41:131–134. doi: 10.1590/0101-60830000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Insel T., Cuthbert B., Garvey M., Heinssen R., Pine D.S., Quinn K. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. The American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 50.Kapur S., Phillips A.G., Insel T.R. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Molecular Psychiatry. 2012;17:1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- 51.Bicikova M., Hill M., Ripova D., Mohr P., Hampl R. Determination of steroid metabolome as a possible tool for laboratory diagnosis of schizophrenia. The Journal of Steroid Biochemistry and Molecular Biology. 2013;133:77–83. doi: 10.1016/j.jsbmb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Maddock J., Zhou A., Cavadino A., Kuzma E., Bao Y., Smart M.C. Vitamin D and cognitive function: a Mendelian randomisation study. Scientific Reports. 2017;7:13230. doi: 10.1038/s41598-017-13189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paternoster L., Tilling K. Davey Smith G. Genetic epidemiology and Mendelian randomization for informing disease therapeutics: conceptual and methodological challenges. PLoS Genetics. 2017;13 doi: 10.1371/journal.pgen.1006944. e1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haycock P.C., Burgess S., Wade K.H., Bowden J., Relton C., Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. The American Journal of Clinical Nutrition. 2016;103:965–978. doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanderWeele T.J., Tchetgen Tchetgen E.J., Cornelis M., Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25:427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess S., Thompson S.G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. American Journal of Epidemiology. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee E.P., Ho J.E., Chen M.H., Shen D., Cheng S., Larson M.G. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metabolism. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim Y.J., Go M.J., Hu C., Hong C.B., Kim Y.K., Lee J.Y. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nature Genetics. 2011;43:990–995. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 59.Heeres R.H., Hoogeveen E.K., Geleijnse J.M., de Goede J., Kromhout D., Giltay E.J. Kidney dysfunction, systemic inflammation and mental well-being in elderly post-myocardial infarction patients. BMC Psychology. 2017;5:1. doi: 10.1186/s40359-016-0170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D'Hooge R., Van de Vijver G., Van Bogaert P.P., Marescau B., Vanholder R., De Deyn P.P. Involvement of voltage- and ligand-gated Ca2+ channels in the neuroexcitatory and synergistic effects of putative uremic neurotoxins. Kidney International. 2003;63:1764–1775. doi: 10.1046/j.1523-1755.2003.00912.x. [DOI] [PubMed] [Google Scholar]
- 61.Kawamura T., Umemura T., Umegaki H., Imamine R., Kawano N., Tanaka C. Effect of renal impairment on cognitive function during a 3-year follow up in elderly patients with type 2 diabetes: association with microinflammation. Journal of Diabetes Investigation. 2014;5:597–605. doi: 10.1111/jdi.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yaffe K., Kurella-Tamura M., Ackerson L., Hoang T.D., Anderson A.H., Duckworth M. Higher levels of cystatin C are associated with worse cognitive function in older adults with chronic kidney disease: the chronic renal insufficiency cohort cognitive study. Journal of the American Geriatrics Society. 2014;62:1623–1629. doi: 10.1111/jgs.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu B., Zheng Y., Alexander D., Manolio T.A., Alonso A., Nettleton J.A. Genome-wide association study of a heart failure related metabolomic profile among African Americans in the Atherosclerosis Risk in Communities (ARIC) study. Genetic Epidemiology. 2013;37:840–845. doi: 10.1002/gepi.21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raffler J., Friedrich N., Arnold M., Kacprowski T., Rueedi R., Altmaier E. Genome-wide association study with targeted and Non-targeted NMR metabolomics identifies 15 novel loci of urinary human metabolic individuality. PLoS Genetics. 2015;11 doi: 10.1371/journal.pgen.1005487. e1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kottgen A., Pattaro C., Boger C.A., Fuchsberger C., Olden M., Glazer N.L. New loci associated with kidney function and chronic kidney disease. Nature Genetics. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chasman D.I., Pare G., Mora S., Hopewell J.C., Peloso G., Clarke R. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000730. e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inouye M., Ripatti S., Kettunen J., Lyytikainen L.P., Oksala N., Laurila P.P. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002907. e1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korostishevsky M., Steves C.J., Malkin I., Spector T., Williams F.M., Livshits G. Genomics and metabolomics of muscular mass in a community-based sample of UK females. European Journal of Human Genetics. 2016;24:277–283. doi: 10.1038/ejhg.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams S.R., Yang Q., Chen F., Liu X., Keene K.L., Jacques P. Genome-wide meta-analysis of homocysteine and methionine metabolism identifies five one carbon metabolism loci and a novel association of ALDH1L1 with ischemic stroke. PLoS Genetics. 2014;10 doi: 10.1371/journal.pgen.1004214. e1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Draisma H.H., Pool R., Kobl M., Jansen R., Petersen A.K., Vaarhorst A.A. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nature Communications. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie W., Wood A.R., Lyssenko V., Weedon M.N., Knowles J.W., Alkayyali S. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62:2141–2150. doi: 10.2337/db12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang T.J., Zhang F., Richards J.B., Kestenbaum B., van Meurs J.B., Berry D. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olden M., Corre T., Hayward C., Toniolo D., Ulivi S., Gasparini P. Common variants in UMOD associate with urinary uromodulin levels: a meta-analysis. Journal of the American Society of Nephrology. 2014;25:1869–1882. doi: 10.1681/ASN.2013070781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kettunen J., Tukiainen T., Sarin A.P., Ortega-Alonso A., Tikkanen E., Lyytikainen L.P. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nature Genetics. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keller M., Schleinitz D., Forster J., Tonjes A., Bottcher Y., Fischer-Rosinsky A. THOC5: a novel gene involved in HDL-cholesterol metabolism. Journal of Lipid Research. 2013;54:3170–3176. doi: 10.1194/jlr.M039420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.