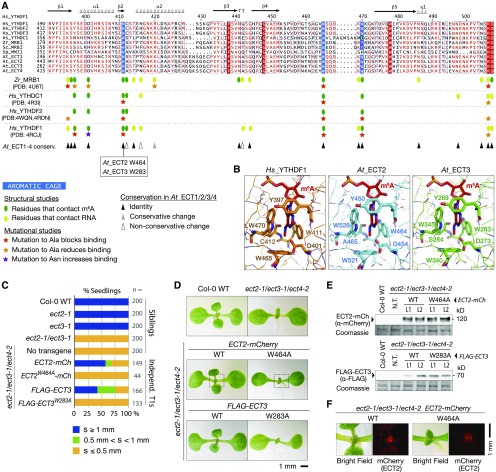
Figure 2.
m6A Binding Sites Are Required for the in Vivo Function of ECT2 and ECT3.
(A) Multiple sequence alignment of part of the YTH domain of the proteins described in Figure 1A. The secondary structure elements of Hs_YTHDF1 are indicated, and amino acids are colored according to level of sequence conservation: red letters, similar residues; red boxes, identical residues (ESPript; Robert and Gouet, 2014). Amino acids that form the methyl-interacting aromatic cage are highlighted in blue. Oval marks and stars summarize the main finding of previous structural and mutational studies of the YTH domain of Zr_MRB1, Hs_YTHDC1, and Hs_YTHDF1/2 as indicated (Luo and Tong, 2014; Xu et al., 2014, 2015; Li et al., 2014b; Zhu et al., 2014). Triangles indicate the degree of conservation of the studied residues in ECT1/2/3/4. PDB, Protein Data Bank (IDs).
(B) Experimentally determined structure of the YTH domain of human YTHDF1 in complex with m6A RNA (PDB: 4RCJ), and models of the YTH domains of ECT2 and ECT3 generated using the homology-modeling server SWISS-MODEL. Residues forming contacts with m6A are highlighted.
(C) Percentages of primary transgenic lines of the indicated genotypes in three categories defined by the length of the first true leaves at 8 d after germination. s, size of first true leaves. Only seedlings with cotyledons longer than 2.5 mm were considered.
(D) Phenotypes of 9-d-old seedlings of the indicated genotypes. Dotted lines delimit the areas analyzed in (F).
(E) Protein blot analyses of wild type and mutant ECT2-mCherry (top) and FLAG-ECT3 (bottom). Two lines (L1 and L2) of each kind with comparable expression levels are shown. Coomassie staining is used as a loading control. N.T., no transgene.
(F) Expression of the wild type and mutant ECT2-mCherry at the shoot apex detected by fluorescence microscopy (mCherry).