Figure 3.
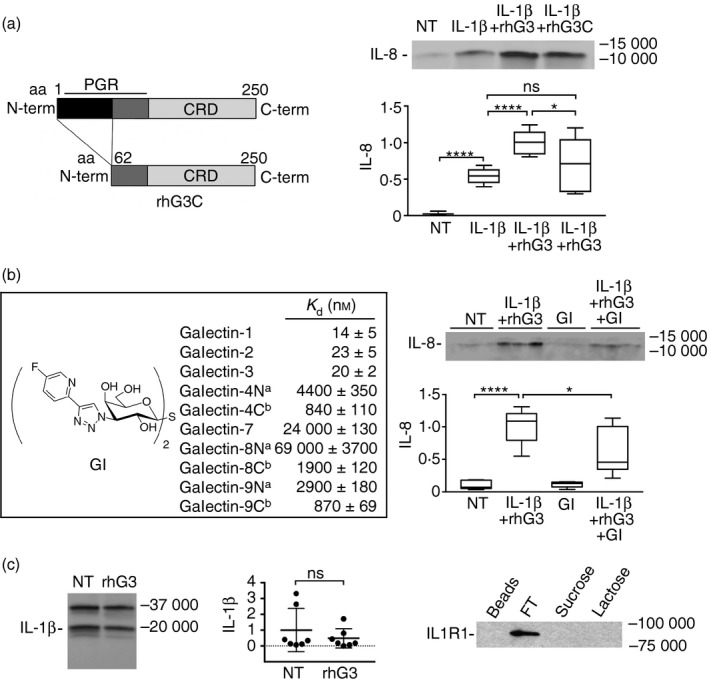
The galectin‐3 pro‐inflammatory activity is established through the N and C termini. (a) A truncated form of galectin‐3 lacking the first 62 amino acids in the N‐terminal domain (rhG3C) was obtained by site‐directed mutagenesis as previously described.15 Stratified cultures of human corneal keratinocytes were incubated with interleukin‐1β (IL‐1β) (10 ng/ml) alone or in combination with 100 μg/ml exogenous full‐length galectin‐3 (rhG3) or rhG3C for 24 hr at 37°. Levels of IL‐8 in the cell‐culture media were determined by immunoblot. CRD, carbohydrate recognition domain; PGR, proline, glycine and tyrosine‐rich domain. NT, not treated. (b) The 1H‐NMR and 13C‐NMR spectra of the galectin inhibitor (GI) are described in the Supplementary material (Figure S1). Galectin affinities were determined in a competitive fluorescence anisotropy assay. Stratified cultures of human corneal keratinocytes were incubated with 1 μm GI alone or in combination with IL‐1β and rhG3 for 24 hr at 37°. Levels of IL‐8 in the cell‐culture media were determined by immunoblot. aN, N‐terminal domain; bC, C‐terminal domain. (c) By immunoblot, addition of exogenous full‐length galectin‐3 had no effect on the levels of IL‐1β in the cell culture media. (d) Cell extracts were collected from stratified keratinocyte cell cultures and subjected to galectin‐3 affinity chromatography. The column was eluted with a non‐competing disaccharide, sucrose, before elution with the competing sugar lactose. FT, flow through. Results in (a) represent three independent experiments performed at least in duplicate, whereas results in (b) represent two independent experiments performed in triplicate. Dissociation constants (K d) in (b) were determined in at least two independent experiments each including at least four data‐points. The box and whisker plots show the 25th and 75th centiles (box), the median and the minimum and maximum data values (whiskers). Results in (c) represent seven independent experiments and data are reported as mean ± standard deviation. Significance was determined using ordinary one‐way analysis of variance with Tukey's post hoc multiple comparison test (a and b), and paired t test (c). *P < 0·05, ****P < 0·0001; ns, non‐significant.
