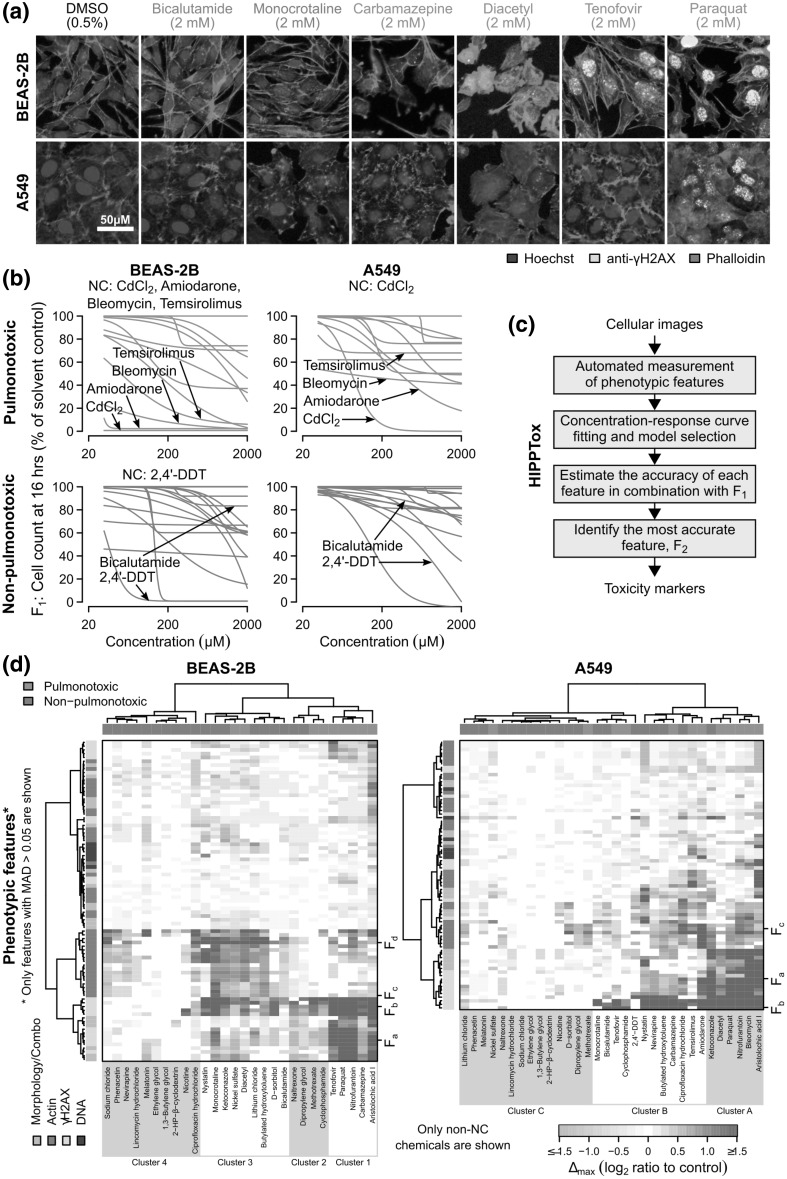
Fig. 2.
Image-based phenotypic profiling shows that BEAS-2B is more active than A549 cells. a Fluorescence microscopy images showing BEAS-2B and A549 cells that had been treated with six of the reference chemicals for 16 h (scale bar = 50 µm). b Percentages of cell count with respect to the solvent controls for all the 30 reference chemicals (red = pulmonotoxic, blue = non-pulmonotoxic reference chemicals). The values were quantified from the cellular images. For BEAS-2B cells, five chemicals, namely, 2,4′-DDT, amiodarone, bleomycin, cadmium(II) chloride (CdCl2), and temsirolimus were found to cause the “no cell” (NC) condition, which is defined to be < 30% cell count at ≥ 125 µM. For A549 cells, only CdCl2 was found to cause NC. c Schematic showing the key steps of HIPPTox used to automatically identify predictive toxicity markers from the obtained cellular images. The same procedures were repeated for both BEAS-2B and A549 cells. d Heatmaps showing changes in the measured phenotypic features of BEAS-2B and A549 cells (“∆max”, see main text) induced by the 33 reference chemicals. Only features with median absolute deviation (MAD) > 0.05 are shown (Fa = mean nuclear γH2AX intensity, Fb = number of γH2AX foci, Fc = total cellular actin intensity, Fd = mean actin object size). The vertical or horizontal groupings of the feature values were based on hierarchical clustering of the columns (chemicals) or rows (features) of the heatmaps, respectively. The Ward’s method (“ward.D” in the hclust() function) was used to agglomerate the clusters. (Color figure online)