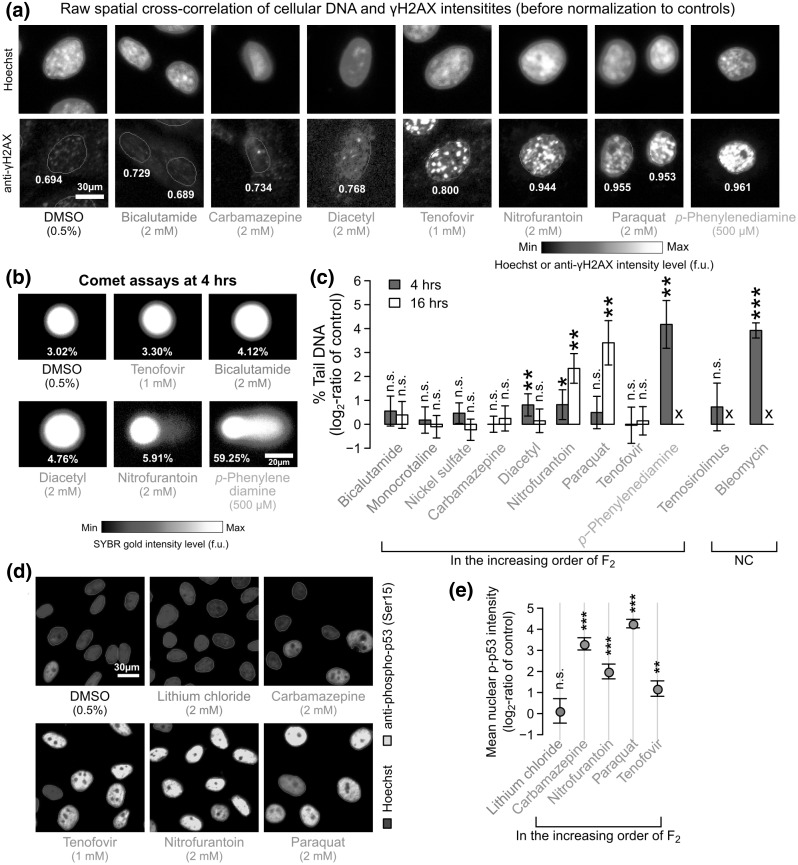
Fig. 4.
Pulmonotoxic chemicals may induce DDR pathway activations dependent or independent of DNA strand breaks. a Fluorescence microscopy images showing the staining patterns of Hoechst and anti-γH2AX in single BEAS-2B cells treated with the indicated chemicals for 16 h. Cells with raw F2 values (indicated below the cells) close to the average raw F2 values across all the cells under the same treatment conditions are shown (scale bar = 30 µm). To allow visual comparisons, all the shown images have been scaled to the same intensity ranges with respect to the solvent controls. b Fluorescence microscopy images showing the DNA spots obtained from the Comet assays of BEAS-2B cells treated with the indicated chemicals for 4 h. DNA spots with % tail DNA values (indicated below the patterns) close to the average % tail DNA values across all the spots obtained under the same treatment conditions are shown (scale bar = 20 µm). To allow visual comparisons, all the shown images have been scaled to the same intensity ranges with respect to the solvent controls. c Changes in the median % tail DNA values obtained from the Comet assays of BEAS-2B cells treated with the indicated chemicals or solvent controls for 4 or 16 h (bars = mean of the log2-ratio values obtained from at least three independent biological replicates, error bars = 95%-tile confidence intervals; X marks = ≤ 5% cells left). d Fluorescence microscopy images showing the staining patterns of Hoechst and anti-phospho-p53 (Ser15) in BEAS-2B cells treated with the indicated chemicals for 16 h. e Changes in the median nuclear phospho-p53 intensity levels of these cells quantified from the images with respect to the solvent controls (dots = mean of the median values, error bars = 95%-tile confidence intervals.) All P values shown in this figure are obtained from two-sided t tests and adjusted for false discovery rates (***P-adjusted ≤ 0.01, **P-adjusted ≤ 0.05, *P-adjusted ≤ 0.10)