ABSTRACT
The heat shock response (HSR) is characterized by the rapid and robust induction of heat shock proteins (HSPs), including HSP70, in response to heat shock and is regulated by heat shock transcription factor 1 (HSF1) in mammalian cells. Poly(ADP-ribose) polymerase 1 (PARP1), which can form a complex with HSF1 through the scaffold protein PARP13, has been suggested to be involved in the HSR. However, its effects on and the regulatory mechanisms of the HSR are not well understood. Here we show that prior to heat shock, the HSF1-PARP13-PARP1 complex binds to the HSP70 promoter. In response to heat shock, activated and auto-PARylated PARP1 dissociates from HSF1-PARP13 and is redistributed throughout the HSP70 locus. Remarkably, chromatin in the HSP70 promoter is initially PARylated at high levels and decondensed, whereas chromatin in the gene body is moderately PARylated afterwards. Activated HSF1 then binds to the promoter efficiently and promotes the HSR. Chromatin PARylation and HSF1 binding to the promoter are also facilitated by the phosphorylation-dependent dissociation of PARP13. Furthermore, the HSR and proteostasis capacity are reduced by pretreatment with genotoxic stresses, which disrupt the ternary complex. These results illuminate one of the priming mechanisms of the HSR that facilitates the binding of HSF1 to DNA during heat shock.
KEYWORDS: HSF1, PARP1, PARP13, chromatin, heat shock, transcription
INTRODUCTION
Cells must maintain an intracellular balance of components, including proteins and nucleic acids, to preserve their health. In order to cope with a variety of environmental and metabolic perturbations, cells have evolved sophisticated surveillance mechanisms, including the proteotoxic stress response, which adjusts proteostasis capacity, or the buffering capacity for misfolded proteins through the regulation of gene expression (1–3). One universally conserved proteotoxic stress response is the heat shock response (HSR), which is characterized by induction of a set of heat shock proteins (HSPs), or chaperones, that facilitate protein folding (4–6).
The HSR is regulated mainly at the level of transcription by heat shock transcription factor (HSF) in eukaryotes. HSF-mediated transcription has been studied intensively using HSP70 as a model gene (7). In Drosophila melanogaster, GAGA-associated factor binds to the HSP70 promoter under unstressed conditions, thereby allowing for the establishment of paused RNA polymerase II (Pol II) and an open chromatin environment that is accessible to HSF (7, 8). In response to heat shock, HSF, which is initially an inactive monomer, is converted to an active trimer and binds to the heat shock response element (HSE) in the HSP70 promoter. It then recruits coactivators and other factors, including Mediator (9), P-TEFb (10), CREB-binding protein (11, 12), and Tip60, which is accompanied by the activation and redistribution of poly(ADP-ribose) polymerase (PARP) throughout the HSP70 locus (13, 14). HSF-dependent recruitment of these coactivators promotes the rapid loss of nucleosomes, the release of stalled Pol II, and the induction of HSP70 transcription.
HSF1 is a master regulator of HSP expression in mammals, whereas all HSF family members (HSF1 to -4) are involved in the regulation of proteostasis capacity via HSP and non-HSP pathways (15, 16). Although GAGA-associated factor is missing in mammalian cells, a small amount of the HSF1 trimer constitutively binds to the HSP70 promoter in complex with replication protein A and the histone chaperone FACT (facilitates chromatin transcription) (17). This complex allows for the establishment of paused Pol II and an open chromatin environment. During heat shock, HSF1 is activated through trimer formation and posttranslational modifications, including phosphorylation (6). Activated HSF1 binds robustly to the HSP70 promoter and dramatically induces its transcription (18, 19) by recruiting various kinds of coactivators, including ASC-2 (20), MLL1 (21), PGC1 (22–24), ATF1 (25), SSBP1 (26), and the SWI/SNF chromatin-remodeling complex, including BRG1 (27, 28). However, it is not clear whether constitutive HSF1 binding and the establishment of paused Pol II are sufficient for efficient HSF1 binding to the HSP70 promoter during heat shock.
PARP1 is a multifunctional regulator of chromatin structure, transcription, and DNA repair (29). We showed previously that HSF1 recruits PARP1 through the scaffold protein PARP13 and that the HSF1-PARP13-PARP1 complex facilitates DNA repair during DNA damage (30). Here we show that the ternary complex binds to the HSP70 promoter under unstressed conditions and that PARP1 is redistributed throughout the HSP70 locus during heat shock. Unexpectedly, heat shock induces the poly(ADP-ribosyl)ation (PARylation) of chromatin in the HSP70 promoter at high levels, as well as in the gene body, which facilitates HSF1 binding to the promoter and promotes the induction of HSP70 expression. Furthermore, DNA damage reduces the HSR and proteostasis capacity by disrupting the formation of the ternary complex.
RESULTS
HSF1-PARP13-PARP1 enhances HSP70 expression during heat shock.
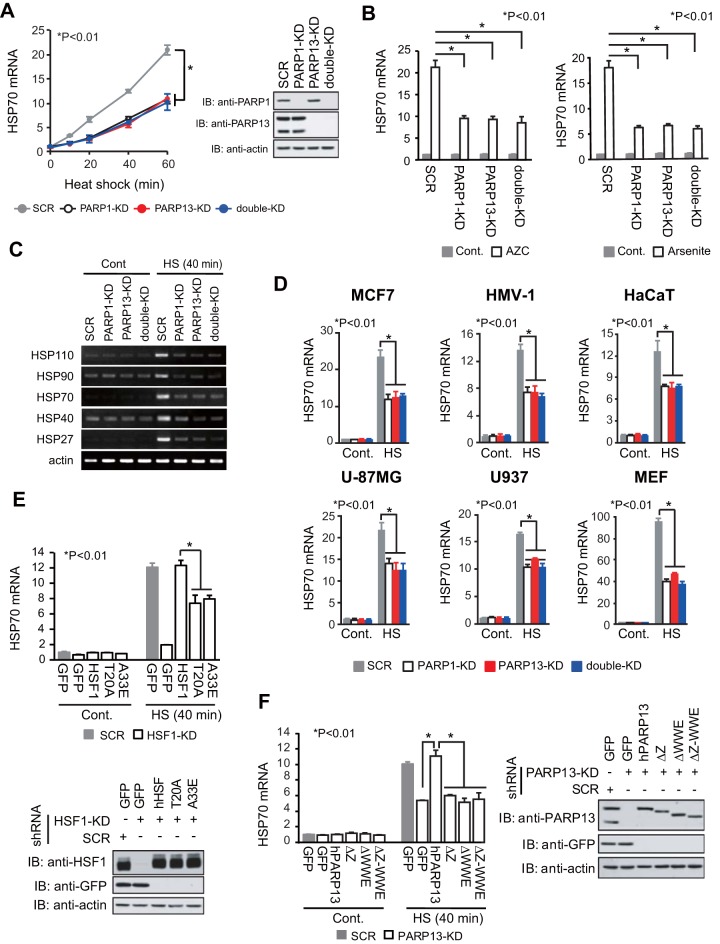
To examine whether the HSF1-PARP13-PARP1 complex regulates the expression of HSP70 (HSPA1A and HSPA1B), we infected HeLa cells with adenoviruses expressing short hairpin RNAs (shRNAs) for PARP1, PARP13, both PARP1 and PARP13 (double knockdown [double-KD]), or scrambled RNA (SCR) as a control. We found that the expression of HSP70 mRNA was induced approximately 20-fold during heat shock at 42°C for 60 min and that the induced levels were markedly lower in PARP1-KD, PARP13-KD, and double-KD cells than in SCR-treated cells at any time point after heat shock (Fig. 1A). The constitutive expression of HSP70 mRNA was unaffected by knockdown of PARP1 and PARP13 (Fig. 1A and B) (30). HSP70 mRNA was also induced by treatment with other proteotoxic stress inducers, including the proline analogue l-azetidine-2-carboxylic acid (AZC) and sodium arsenite, and the induced levels were lower in the same knockdown cells (Fig. 1B). Furthermore, knockdown of PARP1 or PARP13 inhibited the mRNA expression levels of major HSP genes, including HSP110, HSP90, HSP40, and HSP27, during heat shock (Fig. 1C).
FIG 1.
HSF1-PARP13-PARP1 enhances HSP70 expression during heat shock. (A) PARP1, PARP13, or both PARP1 and PARP13 (double-KD) were knocked down by infection of HeLa cells with adenoviruses expressing the corresponding shRNAs. As a control, cells were infected with an adenovirus expressing scrambled RNA (SCR). (Left) The cells were treated with a heat shock at 42°C for the periods indicated, and HSP70 mRNA levels were quantified by RT-qPCR (n = 3). Analysis for statistically significant differences was performed by ANOVA. (Right) Extracts from cells before heat shock treatment were subjected to immunoblotting (IB). Full-length and truncated forms of PARP13 were detected. (B) Cells in which PARP1, PARP13, or both had been knocked down were treated with 5 mM AZC (left) or 20 μM sodium arsenite (right) for 6 h. HSP70 mRNA levels were quantified by RT-qPCR (n = 3). Cont., control. (C) RT-PCR analysis of a set of HSP and β-actin genes was performed using control and heat-shocked (HS) (42°C for 40 min) cells in which PARP1, PARP13, or both (double-KD) had been knocked down. (D) Cells in which PARP1, PARP13, or both had been knocked down were heat shocked (42°C for 40 min). HSP70 mRNA levels were quantified by RT-qPCR (n = 3). (E) Cells in which endogenous HSF1 had been replaced with either GFP, wild-type hHSF1, or mutated hHSF1 were heat shocked (42°C for 40 min). (Top) HSP70 mRNA levels were quantified by RT-qPCR (n = 3). (Bottom) Extracts from cells were subjected to immunoblotting. (F) Cells in which endogenous PARP13 had been replaced with either GFP, wild-type hPARP13, or mutated hPARP13 were heat shocked (42°C for 40 min). (Left) HSP70 mRNA levels were quantified by RT-qPCR (n = 3). (Right) Extracts from cells were subjected to immunoblotting. Analysis for statistically significant differences was performed using Student's t test.
It is controversial whether PARP1 promotes or inhibits the HSR in mammalian cells and tissues during heat shock (31–33). We confirmed that knockdown of PARP1 or PARP13 reduced the induction of HSP70 mRNA in mouse embryonic fibroblasts (MEFs) and various human cell lines, including MCF7, HMV-I, HaCaT, U-87MG, and U937 cells (Fig. 1D). We previously identified human HSF1 (hHSF1) point mutants (hHSF1-T20A, hHSF1-A33E), which cannot interact with PARP13, and hPARP13 deletion mutants (hPARP13-ΔZ, hPARP13-ΔWWE, hPARP13-ΔZ-ΔWWE), which cannot interact with HSF1 (30). To exclude indirect effects of PARP1 knockdown, we replaced endogenous HSF1 with the interaction mutants of hHSF1 and found that HSP70 mRNA expression in these cells was reduced during heat shock (Fig. 1E). Furthermore, we confirmed the reduced expression of HSP70 mRNA during heat shock in cells expressing the hPARP13 interaction mutants (Fig. 1F). These results indicate that the HSF1-PARP13-PARP1 complex enhances HSP70 expression during heat shock.
HSF1-PARP13-PARP1 facilitates the PARylation of chromatin in the HSP70 promoter as well as in the gene body.
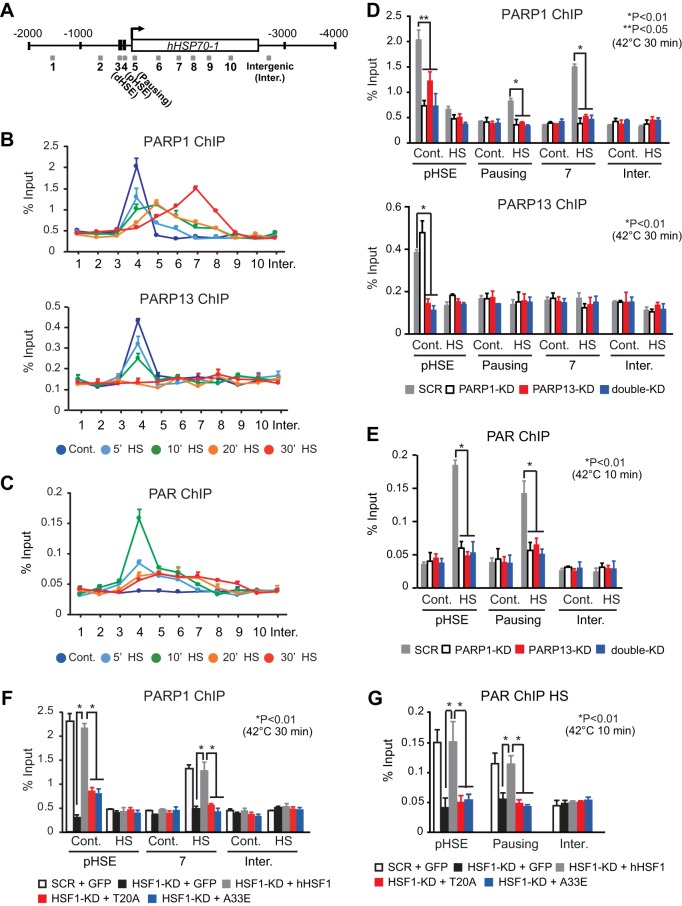
We wanted to understand the mechanisms by which the HSF1-PARP13-PARP1 complex enhances stress-induced HSP70 expression. In Drosophila, PARP is redistributed from the 5′ end of HSP70 to regions throughout the HSP70 locus during heat shock (14). Chromatin immunoprecipitation (ChIP) assays showed that PARP1 and PARP13, like HSF1 (17), were located on the proximal HSE (pHSE) of the HSP70 promoter, but not on the distal HSE (dHSE), under unstressed conditions (Fig. 2A and B). PARP1 and PARP13 mostly disappeared from the pHSE after heat shock at 42°C for 20 min, and only PARP1 was redistributed to the gene body by 30 min (Fig. 2B). It is worth noting that PARP1 was not redistributed to regions upstream of the pHSE. We then examined the PARylation of chromatin by use of an antibody specific for PAR polymers, which was shown previously to detect the PARylation of the Drosophila HSP70 locus (14). Surprisingly, the chromatin was PARylated at an exceptionally high level on the pHSE after heat shock for 10 min and was modestly PARylated on regions 5 and 6 at the same time (Fig. 2C). Subsequently, PARylation levels decreased on the pHSE and increased modestly on regions 7 to 9 within 30 min.
FIG 2.
PARylation of chromatin at the pHSE in the HSP70 promoter is robustly induced during heat shock. (A) Schematic view of the human HSP70-1 locus. Shaded boxes indicate DNA regions amplified by ChIP-qPCR. (B) Occupancy of PARP1 and PARP13 on the HSP70 locus in control HeLa cells (Cont.) and in cells treated with a heat shock (HS) at 42°C for 5, 10, 20, or 30 min. ChIP-qPCR was performed on each of the 10 DNA regions (x axes) indicated in panel A (n = 3). (C) Occupancy of PAR on the HSP70 locus in control cells and in cells treated with a heat shock for the indicated periods (n = 3). (D) Cells in which PARP1, PARP13, or both (double-KD) had been knocked down were either left untreated (Cont.) or treated with a heat shock for 30 min. ChIP-qPCR of PARP1 and PARP13 on the pHSE, pausing region, and region 7 was performed (n = 3). (E) ChIP-qPCR of PAR was performed using control and heat-shocked (10 min) cells in which PARP1, PARP13, or both (double-KD) had been knocked down (n = 3). (F) Cells in which endogenous HSF1 had been replaced with GFP, wild-type hHSF1, or mutated hHSF1 were either left untreated (Cont.) or treated with a heat shock for 30 min. ChIP-qPCR of PARP1 on the pHSE and region 7 was performed (n = 3). (G) ChIP-qPCR of PAR was performed using control and heat-shocked (10 min) cells in which endogenous HSF1 had been replaced with GFP, wild-type hHSF1, or mutated hHSF1 (n = 3). Analysis for statistically significant differences was performed using Student's t test.
We investigated the effects of PARP13 knockdown on PARP1 localization and redistribution during heat shock. We found that PARP1 disappeared from the pHSE with PARP13 knockdown under unstressed conditions, whereas constitutive PARP13 occupancy was not affected by PARP1 knockdown (Fig. 2D). PARP1 was not redistributed to the gene body (pausing region and region 7) during a heat shock with PARP13 knockdown (Fig. 2D). Consistently, chromatin was not PARylated on the HSE and gene body (pausing region) after a heat shock for 10 min with PARP13 knockdown (Fig. 2E). When endogenous HSF1 was replaced with its interaction mutants, PARP1 on the pHSE disappeared under unstressed conditions and was not redistributed to the gene body (region 7) during heat shock (Fig. 2F). Consequently, chromatin was not PARylated when endogenous HSF1 was replaced with its interaction mutants (Fig. 2G). These results indicate that the HSF1-PARP13-PARP1 complex facilitates the PARylation of chromatin in the HSP70 promoter as well as in the gene body during heat shock.
PARP1 and PARP13 facilitate HSF1 binding to the HSP70 promoter during heat shock.
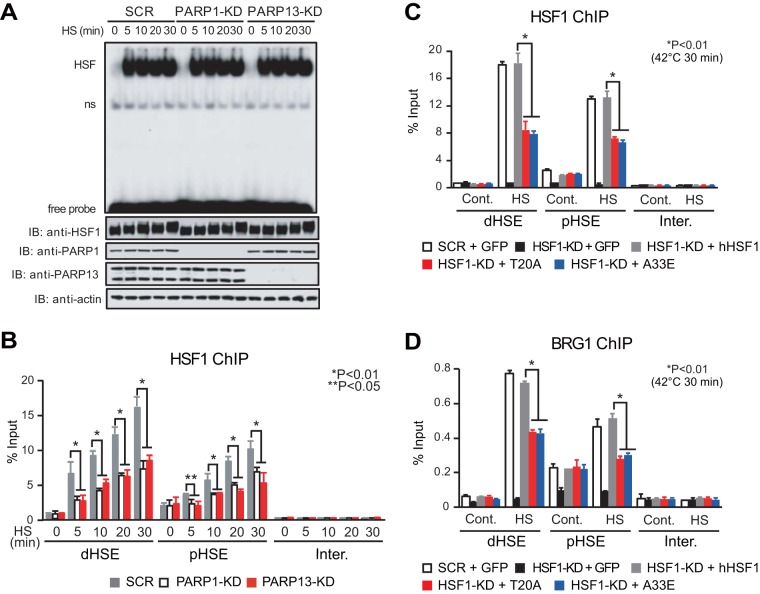
We tested whether the constitutive HSF1-PARP13-PARP1 complex affects HSF1 binding to the HSP70 promoter. We found that knockdown of PARP1 or PARP13 did not effect changes in the degree of hyperphosphorylation and the acquisition of DNA-binding activity by HSF1 during heat shock (Fig. 3A). Remarkably, the knockdown inhibited heat shock-induced HSF1 binding to both the pHSE and dHSE in the HSP70 promoter in vivo (Fig. 3B). Furthermore, replacement of endogenous HSF1 with its interaction mutants inhibited in vivo HSF1 binding to the pHSE and dHSE (Fig. 3C). We then examined the occupancy of BRG1, which is a catalytic component of the chromatin-remodeling complex and is recruited by the C-terminal activation domain of HSF1 (27), on HSEs during heat shock (25). We found that the substitution of HSF1 mutants inhibited BRG1 occupancy during heat shock (Fig. 3D). These results demonstrate that PARP1 and PARP13 facilitate the binding of HSF1 to the HSP70 promoter during heat shock.
FIG 3.
PARP1 and PARP13 facilitate the binding of HSF1 to the HSEs during heat shock. (A) HeLa cells in which PARP1 or PARP13 had been knocked down were treated with a heat shock for the indicated periods. (Top) Whole-cell extracts were prepared and were subjected to an electrophoretic mobility shift assay using a 32P-labeled ideal HSE oligonucleotide. HSF, bands of HSE–HSF1 complex; ns, nonspecific bands; free probe, unbound HSE oligonucleotides. (Bottom) Western blotting (IB) was performed. (B) Cells were treated as described for panel A, and ChIP-qPCR of HSF1 on the pHSE, the dHSE, and an intergenic region (Inter.) was performed (n = 3). (C) Cells in which endogenous HSF1 had been replaced with GFP, wild-type hHSF1, or mutated hHSF1 were heat shocked (42°C for 30 min). ChIP-qPCR of HSF1 was performed (n = 3). (D) Cells were treated, and ChIP-qPCR of BRG1 was performed, as described for panel C (n = 3). Analysis for statistically significant differences was performed using Student's t test.
Heat shock-induced PARP1 activity promotes HSF1 binding to the HSP70 promoter.
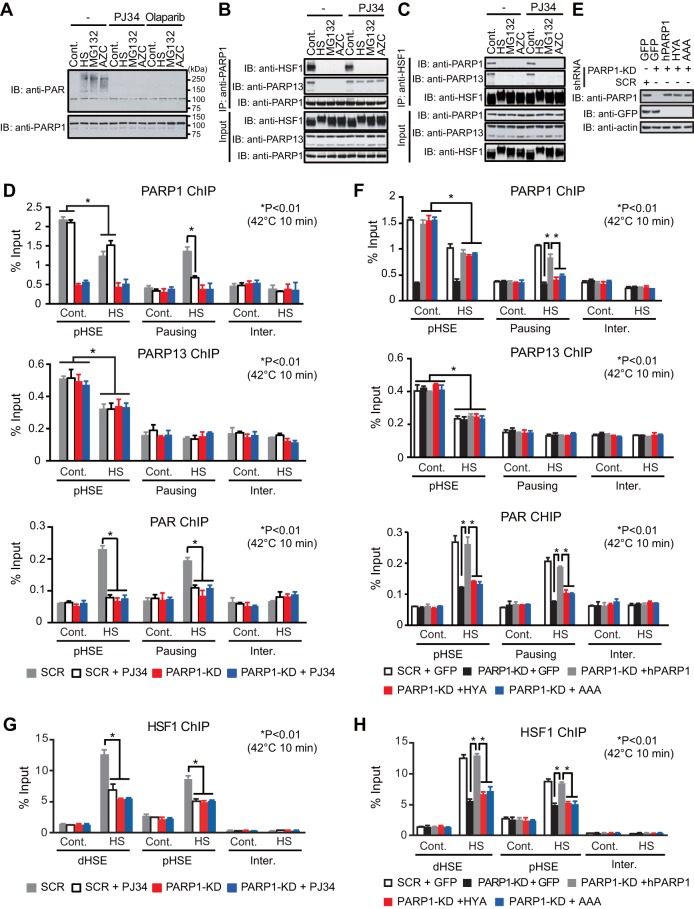
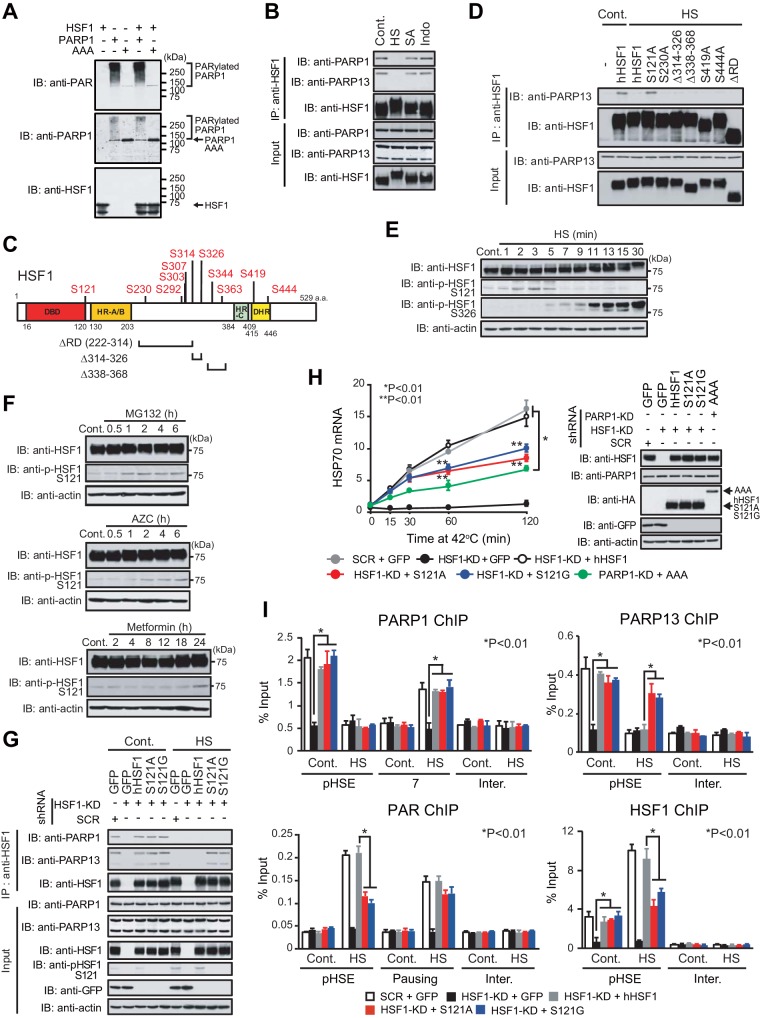
We showed previously that the scaffold protein PARP13 binds directly to both HSF1 and PARP1, and that auto-PARylation of PARP1 causes its dissociation from PARP13 and its redistribution during DNA damage (30). To understand the mechanisms of PARP1 redistribution during proteotoxic stress, we first examined the effects of a heat shock, the proteasome inhibitor MG132, or AZC on ternary complex formation. When the cells were treated with these agents, PARP1 was auto-PARylated and dissociated from PARP13 (Fig. 4A and B). Stress-induced auto-PARylation of PARP1 was inhibited in the presence of the PARylation inhibitor PJ34 or olaparib, a selective inhibitor of PARP1 and PARP2 (PARP1/2) (Fig. 4A). Furthermore, the dissociation of PARP1 was inhibited in the presence of PJ34 (Fig. 4B). On the other hand, PARP13 dissociated from HSF1 in response to proteotoxic stresses even in the presence of PJ34 (Fig. 4C). Thus, heat shock-induced PARP1 activity dissociates PARP1 from PARP13 but does not dissociate PARP13 from HSF1.
FIG 4.
Heat shock-induced PARP1 activity promotes HSF1 binding to the HSP70 promoter. (A) Denatured extracts of HeLa cells treated with either a heat shock at 42°C for 30 min (HS), 10 μM MG132 for 6 h, or 5 mM AZC for 6 h in the presence or absence of PJ34 or olaparib were subjected first to PARP1 immunoprecipitation (IP) and then to immunoblotting (IB) using a PAR antibody. (B) Extracts of HeLa cells treated as described for panel A were subjected first to PARP1 immunoprecipitation and then to immunoblotting. (C) Cells treated as described for panel A were subjected first to HSF1 immunoprecipitation and then to immunoblotting. (D) Cells in which PARP1 had been knocked down were pretreated with PJ34 (20 μM) for 2 h and were then heat shocked at 42°C for 10 min. ChIP-qPCR of PARP1, PARP13, and PAR on the pHSE and pausing regions was performed (n = 3). (E) Extracts of cells in which endogenous PARP1 had been replaced with GFP, wild-type hPARP1, or its inactive mutant (HYA or AAA) were subjected to immunoblotting. (F) Cells were treated as described for panel E, and ChIP-qPCR of PARP1, PARP13, and PAR on the pHSE and pausing region was performed (n = 3). (G) Cells were treated as described for panel D, and ChIP-qPCR of HSF1 on the dHSE and pHSE was performed (n = 3). (H) Cells were treated as described for panel F, and ChIP-qPCR of HSF1 on the dHSE and pHSE was performed (n = 3). Analysis for statistically significant differences was performed using Student's t test.
We next investigated HSF1-mediated recruitment of PARP1 and PARP13 on the pHSE in the HSP70 promoter and the redistribution of PARP1 to the gene body (pausing region). We examined cells treated with a heat shock at 42°C for 10 min, because PARP1 redistribution was easily detectable in the pausing region (Fig. 2B). It was revealed that PARP1 dissociated from the pHSE during a heat shock at 42°C for 10 min even in the presence of PJ34 (Fig. 4D). However, PARP1 was not redistributed to the gene body (pausing region) during a heat shock in the presence of PJ34, probably because PARP1 did not dissociate from PARP13 (Fig. 4B). At the same time, chromatin in the pHSE and gene body was not PARylated (Fig. 4D). We replaced endogenous PARP1 with hPARP1 mutants lacking PARylation activity (HYA and AAA) (Fig. 4E) (30). PARP1 mutants did not redistribute to the gene body (pausing region) during heat shock, although they dissociated from the pHSE (Fig. 4F). In keeping with the fact that the ternary complex facilitates the PARylation of chromatin in the HSP70 promoter (Fig. 2G), treatment of cells with PJ34 or replacement of hPARP1 with mutants lacking PARylation activity markedly suppressed the binding of HSF1 to the HSEs in the HSP70 promoter (Fig. 4G and H). These results indicate that heat shock-induced PARP1 activity is required for PARP1 redistribution and promotes the binding of HSF1 to the HSP70 promoter.
PARP13 dissociation facilitates PARylation and HSF1 binding to the HSP70 promoter.
We wanted to understand the significance and mechanisms of the release of PARP13 from HSF1 in the HSP70 promoter. PARylation is not involved in the release of PARP13, because the stress-induced dissociation was not inhibited in the presence of PJ34 (Fig. 4C). In fact, HSF1 was not PARylated by PARP1, at least in vitro (Fig. 5A). On the other hand, HSF1 is phosphorylated at multiple sites during heat shock but not during treatment with inducers of DNA damage (30), sodium salicylate (34, 35), or indomethacin (36). We found that PARP13 continued to bind to HSF1 in cells treated with sodium salicylate or indomethacin (Fig. 5B), suggesting the involvement of HSF1 phosphorylation in PARP13 dissociation. We generated viral vectors expressing a series of hHSF1 mutants that lacked major heat-inducible phosphorylation residues (Fig. 5C) (37). We then used them to infect HSF1-null MEFs and found that hHSF1-S121A continued to bind to PARP13 during heat shock, whereas wild-type hHSF1 and other mutants dissociated from PARP13 (Fig. 5D).
FIG 5.
PARP13 dissociation facilitates PARylation and HSF1 binding to the HSP70 promoter. (A) hHSF1-His (1 μg) was incubated at 37°C for 30 min with wild-type hPARP1-His (0.1 μg) or mutant hPARP1-His (AAA) (0.1 μg) in the presence of 100 μM NAD. The reaction product was immunoblotted (IB) with an anti-HSF1, anti-PARP1, or anti-PAR antibody. hPARP1 was auto-PARylated, whereas hHSF1 was not PARylated. (B) HeLa cells were treated with either a heat shock (HS) at 42°C for 30 min, 20 mM sodium salicylate for 1 h (SA), or 0.5 mM indomethacin for 1 h (Indo). Cell extracts were prepared and were subjected to HSF1 immunoprecipitation (IP) and immunoblotting. (C) Schematic representation of heat-induced phosphorylation sites in hHSF1 and of hHSF1 mutants lacking these phosphorylation sites. (D) HSF1-null MEFs were infected with an adenovirus expressing wild-type hHSF1 or mutated hHSF1 and were then either left untreated (Cont.) or heat shocked. Cell extracts were prepared and were subjected to HSF1 immunoprecipitation and immunoblotting. (E) HeLa cells were treated with a heat shock at 42°C for the indicated periods, and cell extracts were immunoblotted. (F) HeLa cells were treated with 10 μM MG132, 5 mM AZC, or 10 μM metformin for the indicated periods, and cell extracts were immunoblotted. (G) HeLa cells were first infected with an adenovirus expressing HA-tagged hHSF1, hHSF1-S121A, or hHSF1-S121G and then either left untreated (Cont.) or heat shocked at 42°C for 5 min. Cell extracts were prepared and were subjected to immunoprecipitation and immunoblotting. (H) Cells in which endogenous HSF1 or PARP1 had been replaced with GFP, wild-type hHSF1, hHSF1 mutants (S121A, S121G), or an inactive PARP1 mutant (AAA) were heat shocked at 42°C for the indicated periods. HSP70 mRNA levels were quantified by RT-qPCR (n = 3). Analysis for statistically significant differences was performed using ANOVA (*) or Student's t test (**). (I) Cells in which endogenous HSF1 had been replaced with GFP, wild-type hHSF1, hHSF1-S121A, or hHSF1-S121G were either left untreated (Cont.) or heat shocked for 30 min (PARP1, PARP13, and HSF1 ChIP) or 10 min (PAR ChIP). ChIP-qPCR of PARP1, PARP13, PAR, and HSF1 on the pHSE, pausing region, or region 7 was performed (n = 3). Analysis for statistically significant differences was performed using Student's t test.
We found that phosphorylation of Ser121 was transiently elevated at 2, 3, and 5 min after a heat shock at 42°C in HeLa cells, whereas it was reduced after that time (Fig. 5E) (38, 39). HSF1-Ser121 was also phosphorylated by treatment with MG132 or AZC as well as by the mitochondrial complex I inhibitor metformin (Fig. 5F) (39). We replaced endogenous HSF1 with wild-type hHSF1 or its hHSF1-S121 mutants (hHSF1-S121A, hHSF1-S121G) in HeLa cells and confirmed that the hHSF1-S121 mutants continued to bind to PARP13 during heat shock (Fig. 5G). Expression of HSP70 mRNA in cells expressing hHSF1-S121 mutants was induced 15 and 30 min after a heat shock at levels similar to those in cells expressing wild-type hHSF1 (Fig. 5H). The expression level was further elevated in cells expressing wild-type hHSF1 60 and 120 min after a heat shock, whereas it was hardly elevated in cells expressing mutant HSF1 (Fig. 5H). In marked contrast, the expression levels of HSP70 mRNA were dramatically lower at all time points during heat shock in cells expressing an hPARP1 mutant (Fig. 5H). These results indicate that the dissociation of PARP13 from HSF1 enhances HSP70 expression at later time points after heat shock.
We next examined the occupancy of the HSF1-PARP13-PARP1 complex, and we found that PARP13 stayed on the pHSE in the HSP70 promoter during a heat shock at 42°C for 30 min in cells expressing hHSF1-S121 mutants, whereas PARP1 was redistributed to the gene body (region 7) (Fig. 5I). Remarkably, PARylation of chromatin in the pHSE and HSF1 binding to the pHSE were inhibited in the same cells (Fig. 5I). Thus, PARP13 dissociation facilitates the PARylation of chromatin in the pHSE and the binding of HSF1 to the HSP70 promoter during heat shock.
PARP1 activity promotes the establishment of an active chromatin state.
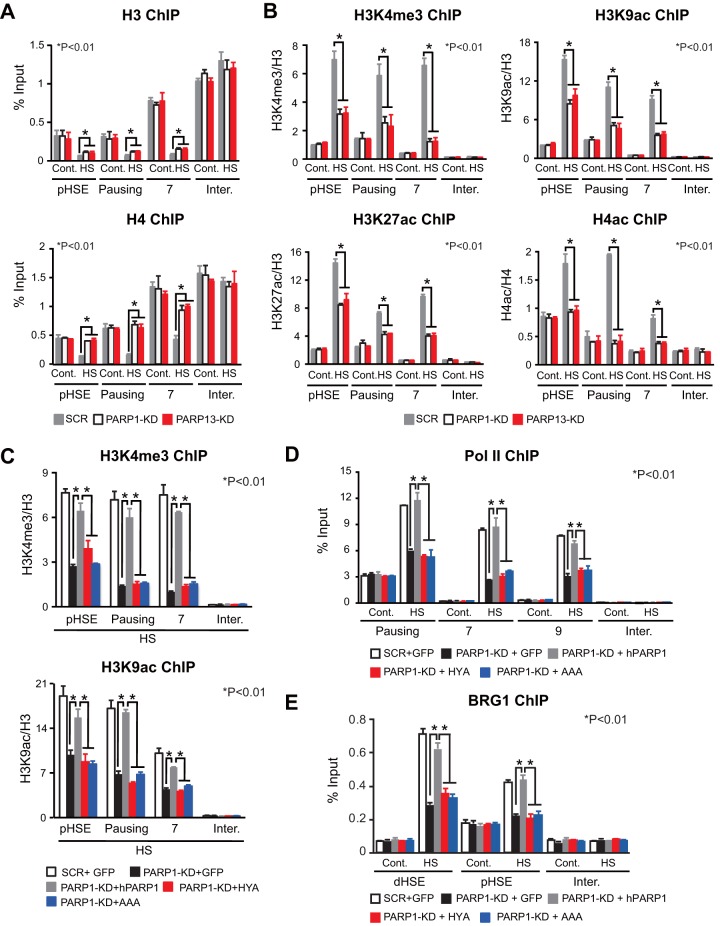
Because the PARylation of histone and nonhistone proteins is associated with chromatin decondensation during heat shock (40, 41), we examined the occupancy of histones H3 and H4. Knockdown of PARP1 or PARP13 did not affect H3 and H4 occupancy on the pHSE and gene body (pausing region and region 7) in the HSP70 locus under unstressed conditions (Fig. 6A). In response to a heat shock, the occupancy of these histones was dramatically reduced on the pHSE and gene body in cells treated with scrambled RNA. In marked contrast, histone H4 occupancy was hardly reduced in PARP1 or PARP13 knockdown cells. Histone H3 occupancy was moderately reduced on the pHSE and gene body in PARP1 or PARP13 knockdown cells during heat shock.
FIG 6.
PARP1 activity promotes the establishment of an active chromatin state. (A) Occupancy of histones H3 and H4 in PARP1 or PARP13 knockdown cells. ChIP-qPCR on the pHSE, pausing region, and region 7 was performed before (Cont.) and after a heat shock (HS) at 42°C for 30 min (n = 3). (B) Levels of active chromatin marks in PARP1 or PARP13 knockdown cells. ChIP-qPCR was performed before and after a heat shock (n = 3). (C) Levels of active chromatin marks in cells in which endogenous PARP1 had been replaced with GFP, wild-type hPARP1, or an inactive mutant of hPARP1 (HYA or AAA). ChIP-qPCR was performed after the heat shock (n = 3). (D) Occupancy of Pol II in cells expressing hPARP1 mutants. ChIP-qPCR on the pausing region and regions 7 and 9 was performed before and after the heat shock at 42°C for 30 min (n = 3). (E) Occupancy of BRG1 in cells expressing hPARP1 mutants. ChIP-qPCR on the pHSE and dHSE was performed (n = 3). Analysis for statistically significant differences was performed using Student's t test.
We then investigated marks of active chromatin, including trimethylation of lysine 4 on histone H3 (H3K4me3), acetylation of lysine 9 on histone H3 (H3K9ac), H3K27ac, and H4ac. These active markers were unaffected by the knockdown of PARP1 or PARP13 under unstressed conditions (Fig. 6B). In response to a heat shock, H4ac levels in both the pHSE and the gene body (pausing region and region 7) were elevated 2- to 4-fold, respectively, in cells treated with scrambled RNA, whereas these levels were hardly elevated in PARP1 or PARP13 knockdown cells. The levels of active marks in histone H3 were markedly elevated in cells treated with scrambled RNA, but they were elevated modestly in PARP1 or PARP13 knockdown cells (Fig. 6B). Furthermore, replacement of endogenous PARP1 with hPARP1 mutants reduced elevated levels of H3K4me3 and H3K9ac on the HSE and gene body during heat shock (Fig. 6C). The same substitution reduced Pol II occupancy on the gene body during heat shock (Fig. 6D) and also BRG1 occupancy on the pHSE and dHSE (Fig. 6E). These results demonstrate that PARP1 activity promotes the establishment of an active chromatin state during heat shock.
DNA damage inhibits the HSR and proteostasis capacity.
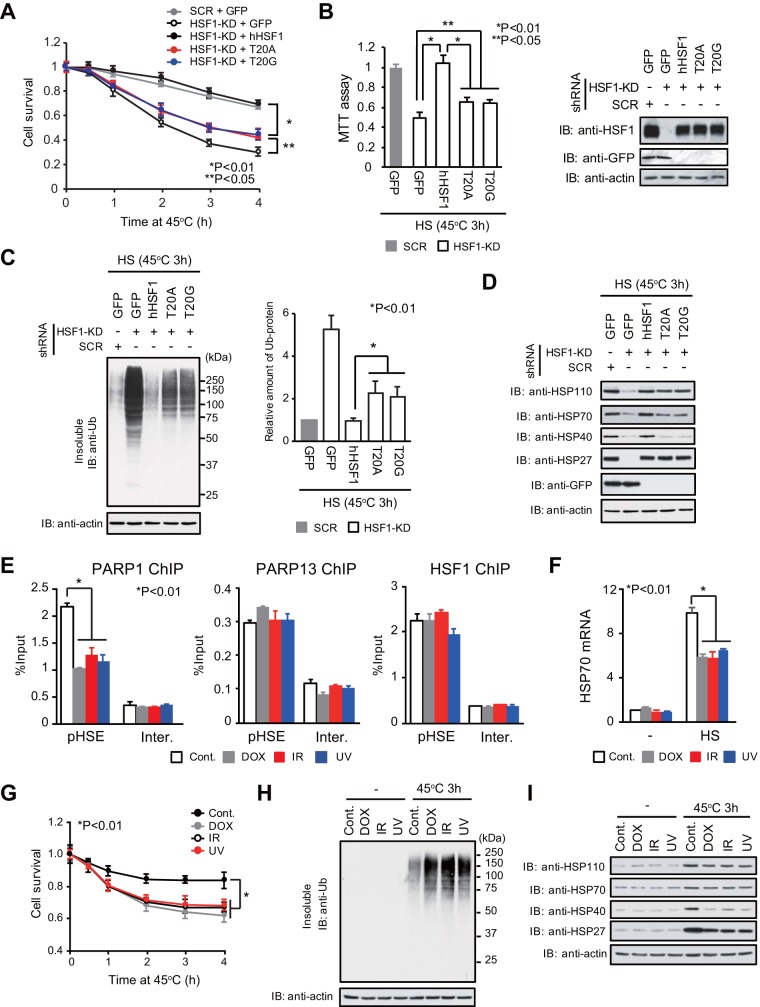
A reduction in HSF1-mediated transcription may be associated with reduced proteostasis capacity and cell survival (25). We showed that cell survival was markedly reduced by replacement of endogenous HSF1 with hHSF1 interaction mutants during an extreme heat shock at 45°C (Fig. 7A and B). The reduction in cell survival was accompanied by elevated accumulation of insoluble ubiquitylated misfolded proteins within the cells (Fig. 7C). In agreement with this finding, the protein levels of HSP110, HSP70, and HSP40 after a heat shock at 45°C for 3 h were lower in cells expressing hHSF1 mutants than in cells expressing wild-type hHSF1 (Fig. 7D). Thus, the HSF1-PARP13-PARP1 complex supports cell survival during heat shock, in part by modulating proteostasis capacity.
FIG 7.
DNA damage inhibits the HSR and proteostasis capacity. (A) HeLa cells in which endogenous HSF1 had been replaced with its mutants were heat shocked at 45°C (HS) for the indicated periods. Viable cells excluding trypan blue were counted (n = 3). Analysis for statistically significant differences was performed by ANOVA. (B) Cells expressing hHSF1 mutants were treated with a heat shock at 45°C for 3 h. (Left) MTT assays were performed (n = 3). (Right) Cell extracts were subjected to immunoblotting (IB). Analysis for statistically significant differences was performed using Student's t test. (C) Cells expressing hHSF1 mutants were heat shocked at 45°C for 3 h. The accumulation of insoluble ubiquitylated proteins was examined by Western blotting using an anti-Ub antibody (left) and was quantified (right) (n = 3). β-Actin levels in the soluble fraction are also shown. Analysis for statistically significant differences was performed using Student's t test. (D) Cells expressing hHSF1 mutants were heat shocked at 45°C for 3 h. Cell extracts were prepared and were subjected to immunoblotting. (E) HeLa cells either were treated with DOX for 6 h or were treated with IR or UV and were allowed to recover for 1 h. Then ChIP-qPCR of PARP1, PARP13, and HSF1 on the pHSE was performed (n = 3). Analysis for statistically significant differences was performed using Student's t test. (F) DNA damage reduced heat shock-induced HSP70 expression. Cells were treated with DOX, IR, or UV as described for panel E. The cells were then heat shocked at 42°C for 3 h, and HSP70 mRNA levels were quantified by RT-qPCR (n = 3). (G) Cells were treated with DOX, IR, or UV as described for panel E. These cells were heat shocked at 45°C for the indicated periods, and viable cells were counted (n = 3). Analysis for statistically significant differences was performed by ANOVA. (H) Cells were treated as described for panel G, and the accumulation of insoluble ubiquitylated proteins was examined by Western blotting. β-Actin levels in the soluble fraction are also shown. (I) Extracts were prepared from cells treated as described for panel G and were subjected to immunoblotting.
Because PARP1 dissociated from the HSF1-PARP13 complex during genotoxic stress (30), we examined the effects of DNA damage. We found that PARP1 was partially released from the pHSE in the HSP70 promoter when the cells either were treated with doxorubicin (DOX) for 6 h or were treated with ionizing radiation (IR) or UV and allowed to recover for 1 h (Fig. 7E). The induction of HSP70 mRNA after a heat shock at 42°C for 3 h was remarkably reduced in cells pretreated with agents causing genotoxic stress (Fig. 7F). Furthermore, cell survival was markedly reduced, and insoluble ubiquitylated misfolded proteins accumulated to high levels, during an extreme heat shock at 45°C when the cells were pretreated with agents causing genotoxic stress (Fig. 7G and H). Protein levels of HSP110, HSP70, HSP40, and HSP27 after heat shock were lower in cells pretreated with genotoxic-stress-causing agents than in untreated cells (Fig. 7I). These results indicate that agents causing genotoxic stress inhibit the induction of HSPs and proteostasis capacity during heat shock by disrupting the HSF1-PARP13-PARP1 complex.
DISCUSSION
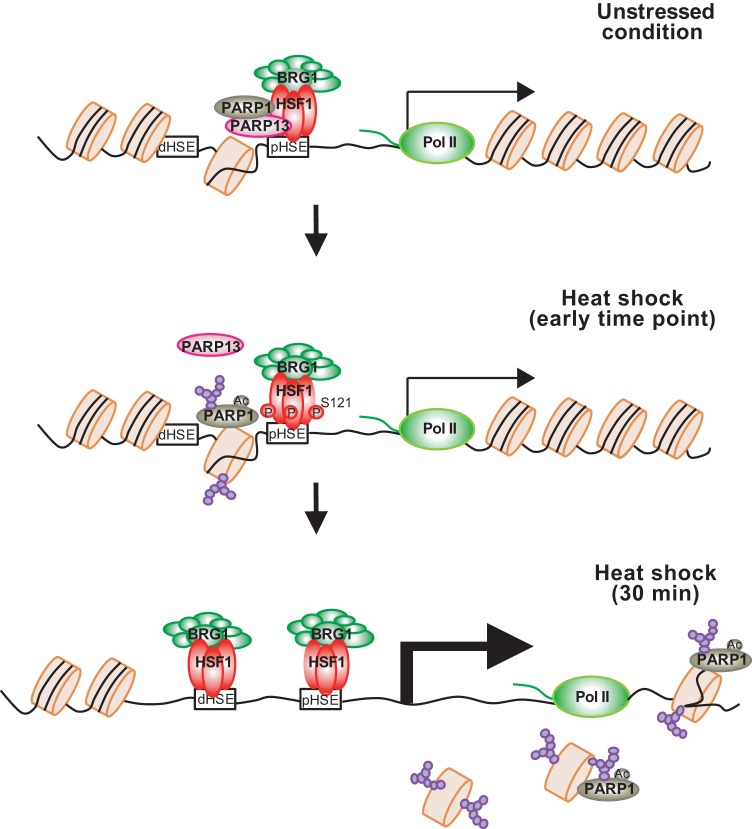
Mammalian HSF1 constitutively binds to the HSP70 promoter in complex with replication protein A and allows for the establishment of paused Pol II at a downstream region of the transcription start site and an open chromatin environment (17). We wondered whether the chromatin environment in the HSP70 locus is further regulated by the HSF1-PARP13-PARP1 complex or not (30). In this study, we demonstrated that the open chromatin environment created by HSF1 binding and paused Pol II is not sufficient for HSF1 to bind efficiently to the HSP70 promoter during heat shock. PARP1 and PARP13 have no effect on the occupancy of H3 and H4 histones, histone modifications, or constitutive HSP70 expression, but they are required for efficient HSF1 binding to the HSP70 promoter, establishment of an active chromatin status, and induction of HSP70 expression during heat shock in mammalian cells (Fig. 8). We propose a priming mechanism of the HSR that facilitates HSF1 binding to DNA during heat shock.
FIG 8.
Regulatory mechanisms of the HSR in mammalian cells. Prior to heat shock, a small amount of HSF1 binds to the pHSE in the HSP70 promoter and allows for the establishment of paused Pol II and an open chromatin environment (unstressed condition). HSF1 also recruits PARP1 through a scaffold protein, PARP13. In response to a heat shock, activated and auto-PARylated PARP1 dissociates from HSF1-PARP13 and is redistributed throughout the HSP70 locus. It PARylates chromatin around the pHSE initially (heat shock, early time point) and around the gene body afterward (heat shock, 30 min). Heat shock-activated HSF1 binds efficiently to the pHSE and dHSE, whose chromatin is decondensed by PARylation, and robustly induces HSP70 transcription. PARP13 also dissociates from HSF1 in a manner dependent on the phosphorylation of HSF1-Ser121.
The roles of PARP in the HSR have been studied in Drosophila melanogaster. Drosophila PARP is required for the formation of puffs containing HSP70 loci and the induction of HSP70 expression (40, 41). PARP binds to the 5′ end of HSP70 (within the first 2 nucleosomes) under unstressed conditions. In response to heat shock, PARP is activated and redistributed throughout HSP70 loci, and PARylation of chromatin increases initially at the 5′ end of HSP70, where PARP binds, and accumulates further downstream afterwards (14). This PARylation promotes chromatin decondensation in the gene body and the induction of HSP70 expression. Unlike Drosophila PARP, human PARP1 is associated with the HSP70 promoter (31) and has been suggested to colocalize with HSF1 on the genome (30). Prior to heat shock, PARP1 occupies the pHSE in the HSP70 promoter by interacting with the HSF1-PARP13 complex (Fig. 2). Heat shock induces auto-PARylation of PARP1, which triggers its dissociation from HSF1-PARP13 and its redistribution throughout HSP70 loci (Fig. 4A to F). Remarkably, chromatin in the pHSE, which is a constitutive PARP1 binding site, is initially PARylated at the highest level within HSP70 loci, whereas chromatin in the gene body is also moderately PARylated afterwards (Fig. 2). As a result, the chromatin status at the pHSE is relaxed (Fig. 6) due to PARylation-mediated chromatin decondensation (14, 42, 43), and heat shock-activated HSF1 binds efficiently to the HSP70 promoter (Fig. 3 and 4). Although the mechanism by which auto-PARylated PARP1 is redistributed along the HSP70 locus is unclear, this modification could enhance its transient interaction with chromatin proteins such as histones at a specific state (44, 45).
Of note, the scaffold protein PARP13 must be released from HSF1 in a manner that is dependent on phosphorylation at HSF1-Ser121 (Fig. 5). HSF1-Ser121 was originally identified as one of the multiple phosphorylation sites in heat-shocked cells (37). Later, it was shown that this residue is constitutively phosphorylated at a low level and is dephosphorylated during heat shock (38, 39). We showed that phosphorylation at HSF1-Ser121 is transiently induced during a heat shock at 42°C (Fig. 5E) and is induced continuously during MG132 or AZC treatment (Fig. 5F). Replacement with hHSF1-S121 mutants blocked the dissociation of HSF1-PARP13 during heat shock and inhibited the PARylation of chromatin on the pHSE in the HSP70 promoter and HSF1 binding (Fig. 5I). Our observations suggest that phosphorylation at Ser121 prior to heat shock by treatment with reagents such as metformin inhibits the HSR (39), in part because of the blockade of ternary complex formation. In contrast to the HSR, PARP13 does not dissociate from HSF1, and chromatin at the GADD34 promoter is not PARylated during DNA damage response (30).
Taking these findings together with our previous work (30), we demonstrate that both proteotoxic and genotoxic stresses regulate the formation and activity of the HSF1–PARP13-PARP1 complex, which plays pivotal roles in the HSR and DNA damage response. These observations suggest that proteotoxic and genotoxic stresses affect each other through the regulation of the ternary complex. Indeed, pretreatment with agents inducing genotoxic stress, including DOX, IR, and UV, dissociated PARP1 from HSF1-PARP13, resulting in a reduction in HSP expression and proteostasis capacity during heat shock (Fig. 7E to I). Our observations reveal one mechanism by which UV pretreatment inhibits the expression of HSP70 without affecting HSF1 DNA-binding activity during heat shock (46) and explain the cooperative effects of combining heat exposure with DNA-damaging anticancer drugs (47, 48). It will be of great interest to discover how this ternary complex contributes to the progression of age-related neurodegenerative diseases and other diseases.
MATERIALS AND METHODS
Plasmids and adenoviral vectors.
To generate expression vectors for hemagglutinin (HA)-tagged hHSF1 (hHSF1-HA), hHSF1-T20A-HA, hHSF1-T20G-HA, hHSF1-A33E-HA, and phosphorylation site mutants, cDNA fragments were generated by PCR-mediated site-directed mutagenesis and were inserted into pShuttle-CMV vectors (Stratagene) at the KpnI/XhoI sites (30). Similarly, cDNA fragments of HA-hPARP13, HA-hPARP13-ΔZ, HA-hPARP13-ΔWWE, and HA-hPARP13-ΔZ-ΔWWE were generated and were inserted into pShuttle-CMV at the BamHI/EcoRI sites, and cDNA fragments of HA-hPARP1, HA-hPARP1-HYA, and HA-hPARP1-AAA were inserted into pShuttle-CMV at the NotI/XhoI sites as described previously (30). The sequences of pShuttle-CMV expression vectors for phosphorylation site mutants of hHSF1 were verified using a 3500 genetic analyzer (Applied Biosystems). Adenovirus expression vectors, including Ad-hHSF1-S20A-HA and Ad-hHSF1-S20G-HA, were generated in accordance with the manufacturer's instructions (Agilent Technologies). To generate adenovirus vectors expressing short hairpin RNAs against human HSF1, PARP1, PARP13 (Ad-hHSF1-KD, etc.), oligonucleotides containing each target sequence (17, 30) were annealed and were inserted into pCR2.1-hU6 at the BamHI/HindIII sites, and then XhoI/HindIII fragments containing hU6-shRNA were inserted into a pShuttle-CMV vector (Stratagene) (49).
Cell cultures and RNA interference.
HeLa cells were maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum (Sigma-Aldrich). Cells were treated with a heat shock at 45°C for various periods or with 5 mM l-azetidine-2-carboxylic acid (AZC; Tokyo Chemical Industry) for 6 h, with 20 μM sodium arsenite (Sigma-Aldrich) for 6 h, with 10 μM MG132 (Sigma-Aldrich) for 6 h, or with 10 μM metformin hydrochloride (Tocris Bioscience) for 24 h. Cells were also treated with 0.5 μM doxorubicin (DOX; Enzo Life Sciences) for 6 h, or they were exposed to 10 Gy ionizing radiation (IR) (MBR-1520R-4; Hitachi Power Solutions) or to 100 J m−2 UV-C (Airtech UV lamp A15436; Ultra-Violet Products UVX radiometer) and were allowed to recover for 1 h. Some cells were pretreated for 2 h with the PARP inhibitor PJ34 at 20 μM (Enzo Life Sciences) or with 1 μM olaparib, a selective inhibitor of PARP1/2 (Selleck Chemicals).
To knock down HSF1, PARP1, or PARP13, HeLa cells were infected with Ad-sh-hHSF1-KD, Ad-sh-hPARP1-KD, or Ad-sh-hPARP13-KD (1 × 107 PFU ml−1) for 2 h and were maintained in normal medium for 70 h. Nucleotide sequences of shRNAs used for gene knockdown are listed in Table S1 in the supplemental material. To replace endogenous HSF1 with exogenous hHSF1 or its phosphorylation site mutants, cells were infected with Ad-sh-hHSF1-KD (1 × 107 PFU ml−1) for 2 h and were maintained in normal medium for 22 h. The cells were then infected with Ad-hHSF1-HA, Ad-hHSF1-S121A-HA, or Ad-hHSF1-S121G-HA (2 × 106 PFU ml−1) for 2 h and were maintained with normal medium for a further 46 h. Replacement of PARP1 or PARP13 with its mutant was performed in the same way.
Western blotting.
Cells were lysed with NP-40 lysis buffer containing 1.0% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), and protease inhibitors (1 μg ml−1 leupeptin, 1 μg ml−1 pepstatin, and 1 mM phenylmethylsulfonyl fluoride). After centrifugation, aliquots of the supernatant were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After transfer to a nitrocellulose membrane using a Trans-Blot transfer cell (Bio-Rad), the membrane was blocked with 5% milk–phosphate-buffered saline (PBS) at room temperature for 1 h. Primary antibodies were diluted in 2% milk–PBS and were incubated at room temperature for 1 h or at 4°C overnight. The following antibodies were used: anti-HSF1 (αmHSF1j or Millipore ABE1044; dilution, 1:1,000), anti-HSF1 Phospho-Ser121 (A8041; dilution, 1:1,000; Assay Biotechnology Company, Inc.), anti-PARP1 (αhPARP1-1, generated at our laboratory; 1:1,000) (30), anti-PARP13 (αhPARP13-1, generated at our laboratory; 1:1,000) (30), anti-PAR (4335-MC-100; Trevigen; 1:1,000), antiubiquitin (P4D1; Santa Cruz; 1:1,000), anti-β-actin (A5441; Sigma-Aldrich; 1:1,000), anti-green fluorescent protein (anti-GFP) (GF200; Nacalai Tesque; 1:1,000), anti-HA (3F10; Roche; 1:1000), anti-HSP110 (αmHSP110a; 1:1,000) (50), anti-HSP70 (W27; Santa Cruz; 1:1,000), anti-HSP40 (αhHSP40a; 1:1,000) (50), and anti-HSP27 (αhHSP27a; 1:1,000) (50). The membrane was washed with PBS for 5 min three times, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (goat anti-rabbit IgG [Cappel 55689; 1:1,000], goat anti-mouse IgG [Cappel 55563; 1:1,000], or goat anti-rat IgG [Jackson 112-035-003; 1:1,000]) in 2% milk–PBS at room temperature for 1 h. To detect HSF1 phosphorylated Ser121 (phospho-Ser121), primary and secondary antibodies were diluted in 3% bovine serum albumin–Tris-buffered saline containing 0.1% Tween 20 (TBS-T). The membrane was washed three times with TBS-T, and chemiluminescent signals from ECL detection reagents (Amersham) were captured on X-ray film (Super RX; Fujifilm).
Assessment of mRNA.
Total RNA was extracted from HeLa cells using TRIzol (Invitrogen), and first-strand cDNA was synthesized using avian myeloblastosis virus reverse transcriptase (AMV-RT) and oligo(dT)20 in accordance with the manufacturer's instructions (Invitrogen). Reverse transcription-PCR (RT-PCR) was performed using the primers summarized in Table S2 in the supplemental material. Real-time quantitative PCR (qPCR) was performed using a StepOnePlus system (Applied Biosystems) with EagleTaq master mix with ROX (Roche) in accordance with the manufacturer's instructions. Primers used for RT-qPCRs are listed in Table S3 in the supplemental material. Relative quantities of mRNAs were normalized against GAPDH mRNA levels. All reactions were performed in triplicate with samples derived from three experiments.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed using a kit in accordance with the manufacturer's instructions (EMD Millipore). The antibodies used for ChIP assays were anti-HSF1 (ABE1044; Millipore), anti-PARP1 (αhPARP1-1) (30), anti-PARP13 (αhPARP13-1) (30), anti-PAR (4335-MC-100; Trevigen), anti-BRG1 (07-478; Millipore), anti-Pol II (CTD4H8; Millipore), anti-H3 (ab1791; Abcam), anti-H4 (ab7311; Abcam), anti-H3K4me3 (ab1012; Abcam), anti-H3K9ac (07-352; Millipore), anti-H3K27ac (ab4729; Abcam), and anti-H4ac (Active Motif 39925). Real-time qPCR of ChIP-enriched DNAs in the HSP70 locus was performed using the primers listed in Table S4 in the supplemental material. The PCR using primers for regions 3 to 10, but not that using primers for regions 1 and 2, amplified both HSPA1A and HSPA1B. The percentage of input was determined by comparing the cycle threshold value of each sample to a standard curve generated from a 5-point serial dilution of genomic input and was normalized to values obtained using normal IgG. IgG-negative control immunoprecipitations for all sites yielded <0.05% input. All reactions were performed in triplicate with samples derived from three experiments.
Electrophoretic mobility shift assay.
Whole-cell extracts were prepared in buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM dithiothreitol). Aliquots of the extracts (10 μg protein) were mixed with a binding mixture containing a 32P-labeled ideal HSE oligonucleotide probe for 20 min at room temperature and were analyzed on 4% native polyacrylamide gels (51).
Immunoprecipitation.
Cells were lysed with NP-40 lysis buffer. After centrifugation, the supernatant was incubated with 5 μl of antiserum (αmHSF1j or αhPARP1-1) or 2 μg of IgG antibody at 4°C for 16 h and was then mixed with 40 μl protein A-Sepharose beads (GE Healthcare) by rotating at 4°C for 1 h. The complexes were washed five times with NP-40 lysis buffer and were subjected to immunoblotting using the antibodies described above. To detect auto-PARylation of PARP1, we performed denaturing immunoprecipitation (30). Briefly, cells (1 × 107) were lysed with 100 μl of denaturing buffer (1% SDS, 5 mM EDTA, 10 mM β-mercaptoethanol) and were heated at 95°C for 10 min. The denatured lysates were mixed with 0.9 ml of radioimmunoprecipitation assay (RIPA) lysis buffer (1.0% NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 0.5% sodium deoxycholate, 0.1% SDS). They were incubated with 5 μl of antiserum for PARP1 (αhPARP1-1) at 4°C for 16 h, mixed with 40 μl of protein A-Sepharose beads at 4°C for 1 h, and subjected to immunoblotting using the PAR antibody described above.
In vitro poly(ADP-ribosyl)ation assay.
Purified recombinant hHSF1-His (1 μg) (17) was incubated at 37°C for 30 min with purified recombinant wild-type hPARP1-His (0.1 μg) or mutant hPARP1-His (AAA) (0.1 μg) (30) in a reaction mixture (50 μl) containing 50 mM Tris-HCl (pH 7.8), 25 mM MgCl2, 1 mM dithiothreitol, 4 μg of nicked calf thymus DNA, and 100 μM β-NAD (Nacalai Tesque, Kyoto, Japan). The reaction was terminated by the addition of SDS-PAGE sample buffer, and reaction products were subjected to immunoblotting with an anti-HSF1, anti-PARP1, or anti-PAR antibody.
Statistical analysis.
Data were analyzed using Student's t test or analysis of variance (ANOVA). Asterisks in figures indicate that differences were significant (P, <0.01 or <0.05). Error bars represent standard deviations for >3 independent experiments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grants 26116720, 15H04706 (to A.N.), 25440010, and 16K07256 (to M.F.), the Uehara Memorial Foundation (to A.N.), and The Yamaguchi University Pump-Priming Program (to A.N.).
M.F. and A.N. designed the project; M.F., R.T., A.K., and P.S. performed the experiments; M.F. and A.N. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00051-18.
REFERENCES
- 1.Wolff S, Weissman JS, Dillin A. 2014. Differential scales of protein quality control. Cell 157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Hipp MS, Park SH, Hartl FU. 2014. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol 24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Labbadia J, Morimoto RI. 2015. The biology of proteostasis in aging and disease. Annu Rev Biochem 84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C. 1995. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 5.Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Pastor R, Burchfiel ET, Thiele DJ. 2018. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. 2010. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harbor Symp Quant Biol 75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. 2016. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev 30:1731–1746. doi: 10.1101/gad.284430.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JM, Werner J, Kim JM, Lis JT, Kim YJ. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell 8:9–19. doi: 10.1016/S1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 10.Lis JT, Mason P, Peng J, Price DH, Werner J. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev 14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 11.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. 2004. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol 6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 12.Boija A, Mahat DB, Zare A, Holmqvist PH, Philip P, Meyers DJ, Cole PA, Lis JT, Stenberg P, Mannervik M. 2017. CBP regulates recruitment and release of promoter-proximal RNA polymerase II. Mol Cell 68:491–503. doi: 10.1016/j.molcel.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zobeck KL, Buckley MS, Zipfel WR, Lis JT. 2010. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40:965–975. doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petesch SJ, Lis JT. 2012. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell 45:64–74. doi: 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto M, Nakai A. 2010. The heat shock factor family and adaptation to proteotoxic stress. FEBS J 277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- 16.Nakai A (ed). 2016. Heat shock factor. Springer, Tokyo, Japan. [Google Scholar]
- 17.Fujimoto M, Takaki E, Takii R, Prakasam R, Hayashida N, Lemura S, Natsume T, Nakai A. 2012. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell 48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. 2016. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell 62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. 2017. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun 8:255. doi: 10.1038/s41467-017-00151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong S, Kim SH, Heo MA, Choi YH, Park MJ, Yoo MA, Kim HD, Kang HS, Cheong J. 2004. Coactivator ASC-2 mediates heat shock factor 1-mediated transactivation dependent on heat shock. FEBS Lett 559:165–170. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Chen J, Yu J, Yang G, Temple E, Harbinski F, Gao H, Wilson C, Pagliarini R, Zhou W. 2014. Identification of mixed lineage leukemia 1 (MLL1) protein as a coactivator of heat shock factor 1 (HSF1) protein in response to heat shock protein 90 (HSP90) inhibition. J Biol Chem 289:18914–18927. doi: 10.1074/jbc.M114.574053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M. 2012. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res 22:1668–1679. doi: 10.1101/gr.127761.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minsky N, Roeder RG. 2015. Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1α. Proc Natl Acad Sci U S A 112:E5669–E5678. doi: 10.1073/pnas.1516219112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Xu L, Alberobello AT, Gavrilova O, Bagattin A, Skarulis M, Liu J, Finkel T, Mueller E. 2015. Celastrol protects against obesity and metabolic dysfunction through activation of a HSF1-PGC1α transcriptional axis. Cell Metab 22:695–708. doi: 10.1016/j.cmet.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Takii R, Fujimoto M, Tan K, Takaki E, Hayashida N, Nakato R, Shirahige K, Nakai A. 2015. ATF1 modulates the heat shock response by regulating the stress-inducible HSF1-transcription complex. Mol Cell Biol 35:11–25. doi: 10.1128/MCB.00754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. 2015. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun 6:6580. doi: 10.1038/ncomms7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. 2001. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol 21:5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev 17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupte R, Liu Z, Kraus WL. 2017. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto M, Takii R, Takaki E, Katiyar A, Nakato R, Shirahige K, Nakai A. 2017. The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun 8:1638. doi: 10.1038/s41467-017-01807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. 2006. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev 20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fossati S, Formentini L, Wang ZQ, Moroni F, Chiarugi A. 2006. Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem Cell Biol 84:703–712. [DOI] [PubMed] [Google Scholar]
- 33.Mota RA, Hernández-Espinosa D, Galbis-Martinez L, Ordoñez A, Miñano A, Parrilla P, Vicente V, Corral J, Yélamos J. 2008. Poly(ADP-ribose) polymerase-1 inhibition increases expression of heat shock proteins and attenuates heat stroke-induced liver injury. Crit Care Med 36:526–534. [DOI] [PubMed] [Google Scholar]
- 34.Jurivich DA, Pachetti C, Qiu L, Welk JF. 1995. Salicylate triggers heat shock factor differently than heat. J Biol Chem 270:24489–24495. [DOI] [PubMed] [Google Scholar]
- 35.Cotto JJ, Kline M, Morimoto RI. 1996. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J Biol Chem 271:3355–3358. [DOI] [PubMed] [Google Scholar]
- 36.Lee BS, Chen J, Angelidis C, Jurivich DA, Morimoto RI. 1995. Pharmacological modulation of heat shock factor 1 by antiinflammatory drugs results in protection against stress-induced cellular damage. Proc Natl Acad Sci U S A 92:7207–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guettouche T, Boellmann F, Lane WS, Voellmy R. 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem 6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Khaleque MA, Zhao MJ, Zhong R, Gaestel M, Calderwood SK. 2006. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. J Biol Chem 281:782–791. doi: 10.1074/jbc.M505822200. [DOI] [PubMed] [Google Scholar]
- 39.Dai S, Tang Z, Cao J, Zhou W, Li H, Sampson S, Dai C. 2015. Suppression of the HSF1-mediated proteotoxic stress response by the metabolic stress sensor AMPK. EMBO J 34:275–293. doi: 10.15252/embj.201489062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tulin A, Spradling A. 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 41.Petesch SJ, Lis JT. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 43.Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A. 2011. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet 7:e1002206. doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotova E, Jarnik M, Tulin AV. 2010. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci U S A 107:6406–6411. doi: 10.1073/pnas.0914152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muthurajan UM, Hepler MR, Hieb AR, Clark NJ, Kramer M, Yao T, Lugen K. 2014. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci U S A 111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu L, Welk JF, Jurivich DA. 1997. Ultraviolet light attenuates heat-inducible gene expression. J Cell Physiol 172:314–322. [DOI] [PubMed] [Google Scholar]
- 47.Overgaard J. 1976. Combined adriamycin and hyperthermia treatment of a murine mammary carcinoma in vivo. Cancer Res 36:3077–3081. [PubMed] [Google Scholar]
- 48.Issels R. 1999. Hyperthermia combined with chemotherapy—biological rationale, clinical application, and treatment results. Onkologie 22:374–381. [Google Scholar]
- 49.Fujimoto M, Hayashida N, Katoh T, Oshima K, Shinkawa T, Prakasama R, Tan K, Inouye S, Takii R, Nakai A. 2010. A novel mouse HSF3 has the potential to activate nonclassical heat-shock genes during heat shock. Mol Biol Cell 21:106–116. doi: 10.1091/mbc.E09-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A. 2005. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem 280:34908–34916. [DOI] [PubMed] [Google Scholar]
- 51.Nakai A, Morimoto RI. 1993. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol 13:1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.