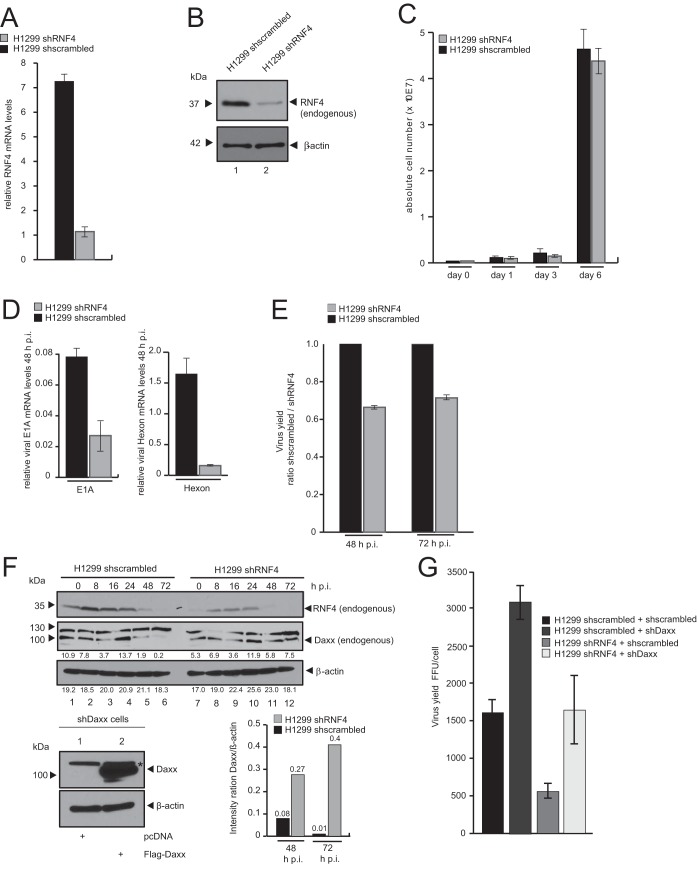
FIG 5.
RNF4 knockdown reduces HAdV viral gene expression and progeny production. (A) H1299 shscrambled and H1299 shRNF4 cells were harvested, and total RNA was extracted, reverse transcribed, and quantified by reverse transcription-PCR (RT-PCR) analysis using primers specific for RNF4. The data were normalized to 18S rRNA levels. The data are presented as relative RNF4 mRNA levels compared between H1299 shRNF4 and H1299 shscrambled control cells. (B) Endogenous RNF4 protein levels in H1299 shscrambled and H1299 shRNF4 cells were determined by preparing whole-cell extracts followed by SDS-PAGE and immunoblotting using RNF4 mouse MAb (kindly provided by T. Urano) and mouse MAb AC-15 (α-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (C) A total of 1 × 105 cells (H1299 shscrambled and H1299 shRNF4) were cultivated, and absolute cell numbers were determined after the indicated time points. The means and standard deviations are presented for three independent experiments. (D) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 20 FFU per cell. The cells were harvested 16 and 48 h postinfection, and total RNA was extracted, reverse transcribed, and quantified by RT-PCR analysis using primers specific for HAdV-C5 E1A and Hexon. The data were normalized to 18S rRNA levels, and the means and standard deviations are presented for three independent experiments. (E) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell. Viral particles were harvested 48 and 72 h posttransfection, and virus yield was determined by quantitative E2A/DBP immunofluorescence staining on HEK293 cells. The means and standard deviations are presented for three independent experiments. Values are shown as the shscrambled/shRNF4 ratio. (F) H1299 shscrambled and H1299 shRNF4 cells were infected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell and harvested after indicated time points postinfection. Total cell extracts were prepared, separated by SDS-PAGE, and subjected to immunoblotting using RNF4 mouse MAb (kindly provided by T. Urano), Daxx rabbit pAb 07-471 (Upstate), and MAb AC-15 (α-β-actin) as a loading control. Daxx and β-actin blots were used for quantitative analysis and amount comparison for Daxx/β-actin intensity ratio at 48 and 72 h p.i. HepaRG shDaxx cells were cotransfected with 5 μg of Flag-Daxx (lower) harvested 24 h posttransfection, and total cell extracts were prepared. Proteins were separated by SDS-PAGE and subjected to immunoblotting. Input levels of total cell lysates were detected using Daxx rabbit pAb 07-471 (Upstate) and mouse MAb AC-15 (α-β-actin) as a loading control. Molecular sizes, in kDa, are indicated on the left, and relevant proteins are on the right. (G) H1299 shscrambled and H1299 shRNF4 cells cotransfected with 5 μg shDaxx construct and 24 h later superinfected with wt virus (H5pg4100) at a multiplicity of 50 FFU per cell. Viral particles were harvested 48 h posttransfection, and virus yield was determined by quantitative E2A/DBP immunofluorescence staining on HEK293 cells. The means and standard deviations are presented for three independent experiments.