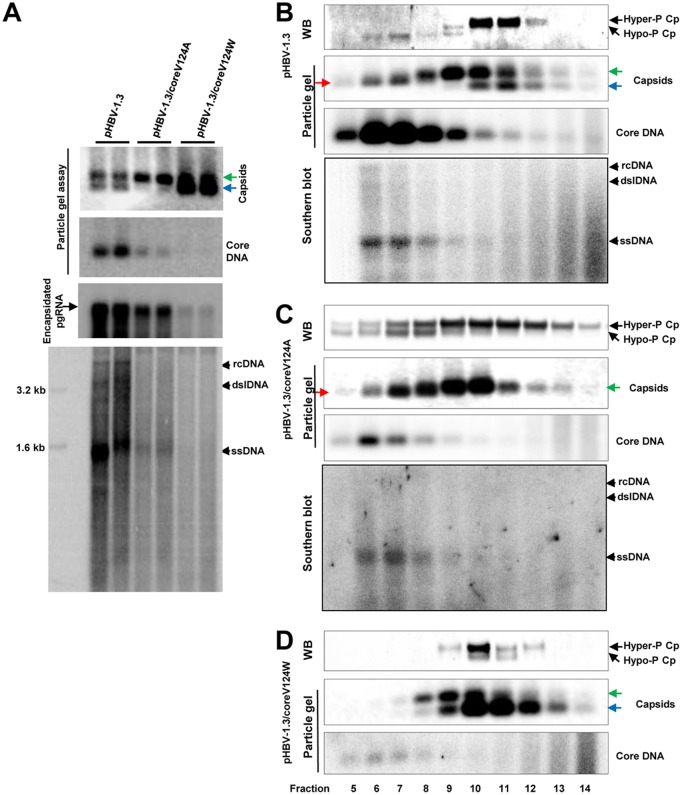
FIG 8.
Analyses of effects of core protein mutation on capsid assembly, DNA replication, and core protein phosphorylation. (A) HepG2 cells were transfected with plasmid pHBV-1.3, pHBV-1.3/coreV124A, or pHBV-1.3/coreV124W and harvested at 72 h posttransfection. (Top and middle) HBV capsids and HBV DNA were detected by a 1.5% native agarose gel assay. The slow- and fast-migrating capsids are indicated by green and blue arrows, respectively. Encapsidated pgRNA was determined by Northern blotting hybridization. (Bottom) The cytoplasmic HBV DNA replication intermediates were analyzed by Southern blotting hybridization. (B to D) HepG2 cells were transfected with plasmid pHBV-1.3 (B), pHBV-1.3/coreV124A (C), or pHBV-1.3/coreV124W (D) and harvested at 72 h posttransfection. Intracellular capsids were sedimented on a sucrose gradient (15% to 30%). Sucrose density, HBV core protein, and DNA from each of the fractions were determined. Core protein and capsids in fractions 5 (high sucrose density) to 14 (low sucrose density) were analyzed by Western blotting and particle gel assays as described in the legend to Fig. 4. HBV DNA replicative intermediates were analyzed by Southern blotting hybridization. The slow- and fast-migrating capsids are indicated by green and blue arrows, respectively. The capsids migrating between the slow and fast capsids are indicated by red arrows.