Figure 5.
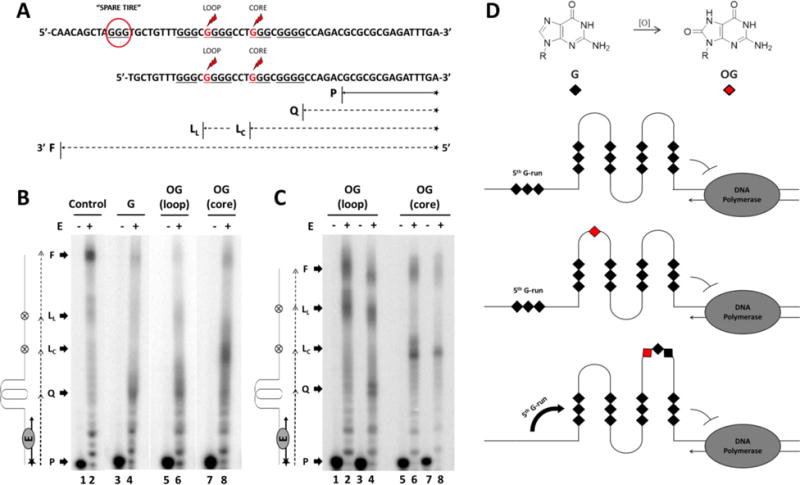
(A) The NEIL3 gene promoter sequence specifying where the “spare tire” domain (enclosed in a red circle) and the OG positions (indicated by the red Gs marked with the red lightning symbols) are placed. Extension of the annealed 15-mer primer P with expected arrest sites either at the G4 site Q, or at the base lesion OG in the loop LL or core position LC are marked while the fully synthesized DNA product is designated as F. (B-C) Autoradiographs of the DNA polymerase stop assay. Primer annealed-template DNAs were incubated at 37 °C for 30 minutes with (+) or without (−) the Klenow Fragment E. (B) DNA polymerase stop assays of five-track NEIL3 G4s with or without OG. Control lanes were reactions using the native strand in LiCl. (C) DNA polymerase stop assays of OG-containing four-track NEIL3 G4s. Lanes 1-2 and 5-6 were control experiments run in buffer with LiCl. (D) Model illustrating the NEIL3 G4 stalling the DNA polymerase and the 5th G-run acting as a “spare tire” that reconstructs the G4 fold in the presence of an oxidative base lesion.
