Figure 4.
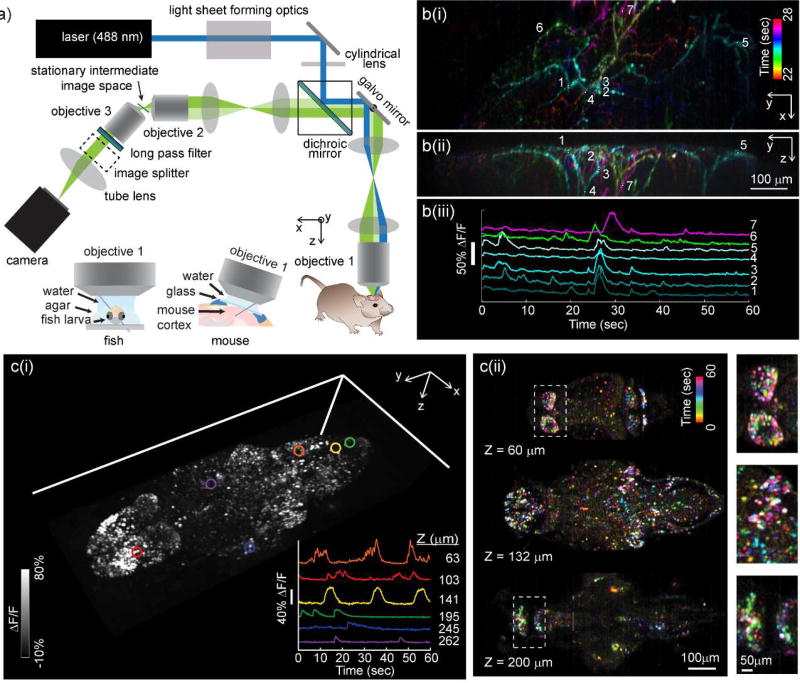
SCAPE microscopy for high-speed 3D neuroimaging. a) Optical layout of a typical SCAPE system permitting oblique sheet scanning through a stationary, single objective lens. Insets show imaging geometries for brain imaging in zebrafish larvae (left) and awake, behaving mouse brain (right). b(i) shows SCAPE data acquired at 10 VPS (140 × 750 × 149 x-y-z voxels = 372 × 1032 × 174 micron field of view (FOV)) in an awake, behaving mouse capturing spontaneous activity in apical dendrites of layer 5 neurons in whisker barrel cortex via GCaMP6f (AAV9.Syn.GCaMP6f) using methods similar to [26], but with improved resolution and SNR compared to our first demonstration. Maximum intensity views from the top (x-y) and side (y-z) are shown for activity occurring between 22 and 28 seconds (with colors denoting time). Raw fluorescence time-courses from the regions of interest indicated are shown as raw data, showing excellent SNR, minimal photobleaching over 60 seconds and the ability to probe firing dynamics along individual dendrites during a single spontaneous event. A time-color merge of all dendritic activity for the full 60 seconds is shown in supplemental figure 3. c) shows imaging of spontaneous activity in the whole brain of larval zebrafish: Data was acquired at 6 VPS over a 820 × 380 × 260 µm FOV (i) shows a volume rendering of a time-maximum intensity projection taken over all 360 time points over a 1 minute run. Inset shows time series extracted from 6 neurons at 6 depth planes within the fish as indicated. (ii) Shows a time-encoded color projection of 3 depth planes showing spontaneous activity over a range of brain regions. Insets show ~2× close ups of indicated regions. 7 day past fertilization HuC:H2B-GCaMP6f fish obtained from Janelia Farm.