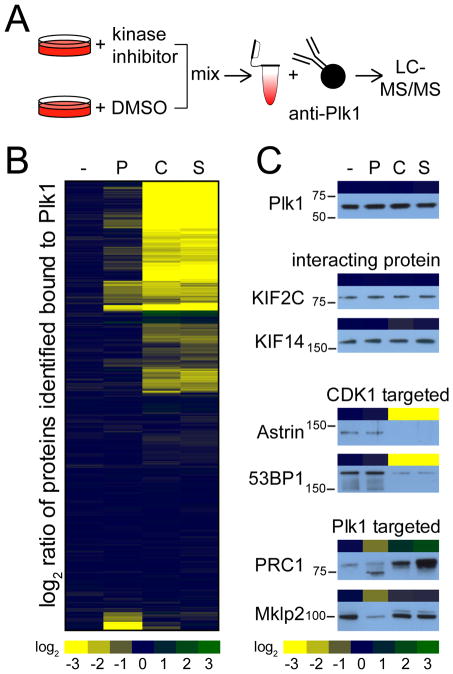
Figure 1. Kinase activity-dependent Plk1 interactome.
(A) Experimental design to identify kinase activity-dependent Plk1 interactors by SILAC. Heavy- and light-labeled HeLa cells were arrested in mitosis and treated with MG132. Heavy-labeled cells were treated with a specified kinase inhibitor or DMSO. Heavy- and light-labeled cells were mixed, lysed, and then Plk1 and its interacting proteins were immunoprecipitated from the lysates, resolved by gel electrophoresis and analyzed by LC-MS/MS. (B) Hierarchical clustering of quantitative differences in the binding of Plk1-interacting proteins in HeLa cells arrested with Taxol (-) or treated with Plk1 inhibitor (“P”; BI2536), Cdk inhibitor (“C”; flavopiridol), or staurosporine (S). (C) Comparison between quantitative mass spectrometry data (heatmap) with Western blot analyses of immunoprecipitated Plk1, the two stable interacting proteins KIF2C and KIF14, the two CDK1 kinase activity-dependent Plk1-interacting proteins Astrin and 53BP1, and the two Plk1 kinase activity-dependent Plk1-interacting proteins Prc1 and Mklp2 upon differential kinase inhibitor treatment.