Abstract
As precision medicine becomes increasingly relevant in healthcare, the field of pharmacogenomics (PGx) also continues to gain prominence in the clinical setting. Leading institutions have begun to implement PGx testing and the amount of published PGx literature increases yearly. The Pharmacogenomics Knowledgebase (PharmGKB; http://www.pharmgkb.org) is one of the foremost worldwide resources for PGx knowledge, and the organization has been adapting and refocusing its mission along with the current revolution in genomic medicine. The PharmGKB website provides a diverse array of PGx information, from annotations of the primary literature to guidelines for adjusting drug treatment based on genetic information. It is freely available and accessible to everyone from researchers to clinicians to everyday citizens. PharmGKB was found over 17 years ago, but continues to be a vital resource for the entire PGx community and the general public.
This article is categorized under:
Translational, Genomic, and Systems Medicine > Translational Medicine
Keywords: pharmacodynamics, pharmacogenetics, pharmacogenomics, pharmacokinetics, pharmGKB
1. INTRODUCTION
President Barack Obama announced in his 2015 State of the Union address the launch of the Precision Medicine Initiative, an effort aimed at advancing the ability of medical professionals to provide individualized care (The Precision Medicine Initiative, 2015). This public statement of support from the White House for precision medicine is just one recent example of how the field has started entering both mainstream healthcare and the public consciousness. Individual companies such as 23andMe and Veritas have begun to engage citizens worldwide in learning more about their genomic information in the context of healthcare. This interest is compounded by the current enthusiasm for ancestral genomic information, such as that offered through http://ancestry.com or National Geographic's Genographic Project. With global initiatives, as well as an increasingly attentive population, precision medicine will rapidly develop in the coming years.
Precision medicine—also known as personalized medicine or genomic medicine—encompasses the way in which individuals develop diseases and respond to treatments differently. Variations within genes can increase or decrease susceptibility to certain diseases. They can also affect how a patient responds to a certain medication, an area of genomic medicine known as pharmacogenomics (Goodman, Cassio, & Livingstone, 2013). While precision medicine mainly focuses on genetic changes, the term also includes factors such as the environment and the microbiome, which also affect disease risk and medication response (The Precision Medicine Initiative, 2015). Pharmacogenomics plays a major role in precision medicine—it has been used successfully in clinical settings (Dunnenberger et al., 2015; Luzum et al., 2017), and its advancement is hampered by fewer ethical considerations as compared to disease genetics (Gershon, Alliey‐Rodriguez, & Grennan, 2014). While the origin of modern pharmacogenetics can be traced back over 50 years (Relling & Evans, 2015; Weinshilboum & Wang, 2006), significant progress in the field has been made within the last decade alone. Advances in genomic sequencing have enabled scientists to more easily identify genomic variation associated with drug‐response phenotypes, and published pharmacogenetics‐related literature has been increasing accordingly (Scott, 2011). Hospitals and universities such as St. Jude Children's Research Hospital, the University of Florida, Vanderbilt University and the Mayo Clinic have been forerunners in implementation of pharmacogenetics in the clinic. Organizations such as the Clinical Pharmacogenetics Implementation Consortium (CPIC, Relling & Klein, 2011) and Dutch Pharmacogenetics Working Group (DPWG, Swen et al., 2008) have greatly assisted in the implementation process through the development and publication of freely available, peer‐reviewed gene–drug dosing guidelines. Furthermore, a growing number of drug labels approved by the US Food and Drug Administration (FDA) now include pharmacogenomic material.
The Pharmacogenomics Knowledgebase (PharmGKB; http://www.pharmgkb.org) is a publicly available resource funded by National Institutes of Health (NIH) and National Institute of General Medical Sciences (NIGMS). Established in 2000, PharmGKB today is the preeminent worldwide resource for pharmacogenomic information, and has been heavily involved in advancing the field through its creation and maintenance of a knowledge base and its work within the NIH‐sponsored Pharmacogenomics Research Network (PGRN). The PharmGKB's role in the community is more critical than ever—the amount of published pharmacogenomic literature steadily increases every year, clinical implementation continues to grow, drug labels from around the globe have begun to include pharmacogenomic data, and next generation sequencing (NGS) has created new opportunities for analyzing the whole genome. The PharmGKB team continues to expand their translational science efforts from the basic science to the bedside, while maintaining a knowledge resource that includes a robust knowledgebase, API and website that is user‐friendly and adaptable. The user interface for the website provides accessibility for everyone from citizens to clinicians to researchers.
2. BASIC PHARMACOGENOMICS AND PHARMGKB
While the terms “pharmacogenetics” and “pharmacogenomics” are technically distinct—pharmacogenetics refers to the study of genetic influence on drug response when only one or several genes are involved, while pharmacogenomics considers variation across the genome—they are often used interchangeably; for brevity, we will use the term PGx within this review. Genetic variation plays a role in drug response in two main ways—affecting how well the drug works (efficacy) and affecting whether the drug causes adverse events (toxicity). Fine‐tuning of efficacy and toxicity is the main goal of PGx implementation in health care. Genetic variation affects efficacy and toxicity mainly through the alteration of a drug's pharmacokinetics or pharmacodynamics.
2.1. Pharmacokinetics
Pharmacokinetics encompasses a drug's absorption, distribution, metabolism and excretion by the body (Ratain & Plunkett Jr, 2003). Proteins are involved in each one of these processes and variations in the genes encoding these proteins can alter how efficiently a drug is metabolized. If a drug is metabolized too quickly it may not be as efficacious or, in the case of prodrugs, the patient may receive a greater amount of the active compound than intended. Alternatively, if the drug is not cleared from the body properly, it may build up and cause toxic side effects.
One example of the way in which genetic changes can affect a drug's pharmacokinetics leading to toxic side effects is the involvement of TPMT variants with the clearance of thiopurine drugs. Thiopurines, which include mercaptopurine, thioguanine, and azathioprine (a prodrug for mercaptopurine), are antineoplastic and immunosuppressive agents used to treat a variety of cancers, as well as inflammatory bowel diseases (Dean, 2012). While all thiopurines are intrinsically toxic, some patients suffer severe or even fatal bone‐marrow toxicity when receiving the drugs, even at standard doses (Abbott, 2003). Beginning in the mid‐1990s, scientists began to discover that the cause of this toxic reaction was due to variations in the gene coding for the enzyme thiopurine S‐methyltransferase (TPMT). Because of its clinical importance, TPMT was the first gene selected by the FDA for public hearings on whether it should be included in drug labeling, and clinical testing for the gene is widely available and applied (Weinshilboum & Wang, 2006).
As shown in the PharmGKB pathway for thiopurine drugs (Zaza et al., 2010, Figure 1), as well as discussed in the PharmGKB review of the TPMT gene (Wang et al., 2010), mercaptopurine and thioguanine exert their cytotoxic effects through the formation of thioguanine nucleotides (TGNs), active metabolites that incorporate into DNA. The TPMT enzyme is responsible for the inactivation of these drugs. Specific variations within the TPMT gene are known to lead to an inactive gene—the most common of these variants are known as TPMT*2, TPMT*3A, and TPMT*3C. However, there are over 40 known TPMT variants defined by the TPMT nomenclature committee (https://www.imh.liu.se/tpmtalleles Linköping University, Institutionen för Medicin Och Hälsa, 2013). The functional status of many of these variants and details on their genetic locations can be found in the gene‐specific information table for TPMT on the PharmGKB website (https://www.pharmgkb.org/page/tpmtRefMaterials). Patients who carry one of the inactive variants have reduced TPMT activity, while patients who carry two inactive variants are TPMT deficient (Relling et al., 2011). Individuals who are TPMT deficient cannot inactivate mercaptopurine and thioguanine, and the drugs instead anabolize into TGNs. This causes a toxic build‐up of TGNs and potentially fatal myelosuppression in 100% of cases. Individuals who carry only one nonfunctional TPMT allele are still at an increased risk of myelosuppression (Relling et al., 2011).
Figure 1.
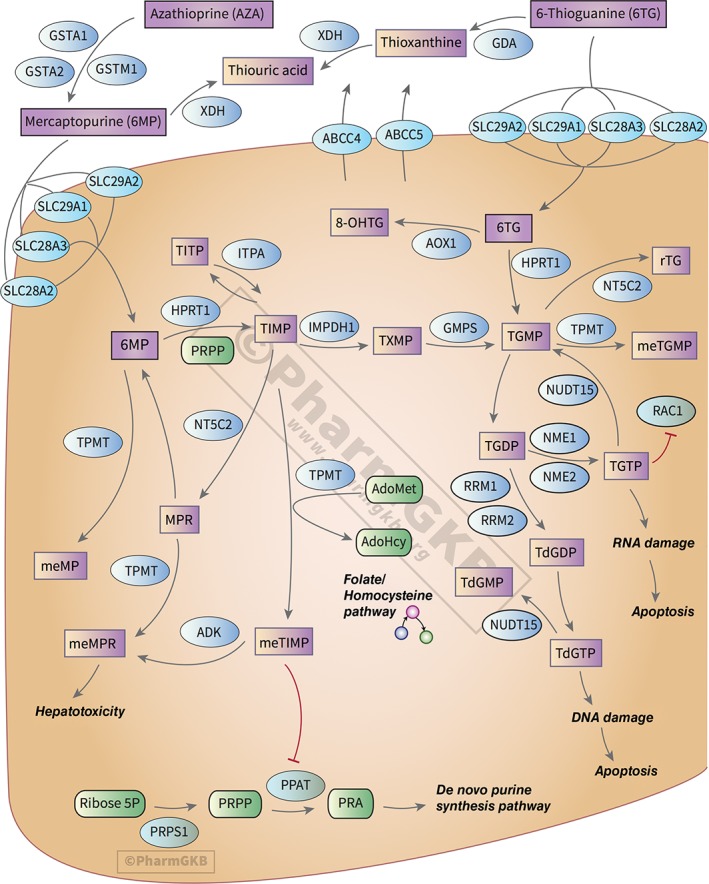
The Pharmacogenomics Knowledgebase (PharmGKB) pathway diagram for thiopurines. PharmGKB pathways detail the pharmacokinetics or pharmacodynamics of pharmacogenomically important drugs, and include text providing detailed background and pharmacogenomic information. Most pathways are created in collaboration with experts in the field, and all pathways are available for download in multiple formats. An interactive version of this pathway can be found on the PharmGKB website at https://www.pharmgkb.org/pathway/PA2040
In addition to providing a detailed illustrated pathway for thiopurine drugs and a detailed review of the TPMT gene, PharmGKB provides pathways for over 100 other drugs and reviews for over 60 genes important to pharmacogenomics. PharmGKB drug‐centric pathways include detailed illustrations showing the pharmacokinetics or pharmacodynamics of a PGx‐relevant drug, as well as text that provides background on the drug, information on its metabolism or mechanism of action, and a detailed review of its pharmacogenomics. Connections on the pathway are supported by literature evidence, and the pathways are available for download in multiple formats, including BioPax. Gene‐based review articles on PharmGKB are referred to as very important pharmacogene (VIP) summaries, and include background information on the gene, as well as an in‐depth discussion of its PGx associations. Figure 2 shows the PharmGKB page for TPMT, where the VIP summary and thiopurine pathway can be accessed, as well as other information about the gene, such as dosing guidelines from CPIC (Relling et al., 2013), variant annotations, and clinical annotations; these data types are discussed in detail in later sections of this review, as well as in Box 1.
Figure 2.
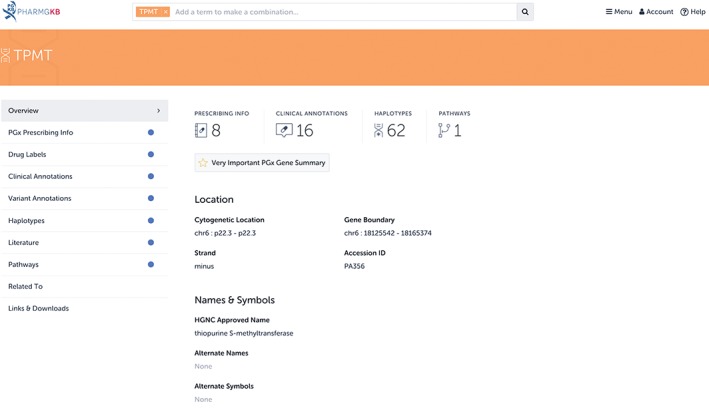
The page for the TPMT gene on the Pharmacogenomics Knowledgebase (PharmGKB). Any information PharmGKB has on the TPMT gene or its variants is included on this page. This includes pathways that the TPMT gene appears in, such as the PharmGKB thiopurine pathway(Zaza et al., 2010) or the very important pharmacogene (VIP) summary article on the gene(Wang et al., 2010). Other types of data that discuss TPMT are also present on this page, such as Food and Drug Administration (FDA)‐approved drug labels or PharmGKB clinical or variant annotations. These data types are discussed in later sections of this review
BOX 1. TYPES OF INFORMATION AVAILABLE ON PharmGKB.
PharmGKB provides multiple types of PGx information for the drugs, genes, genetic variants, and phenotypes it has on the website. These are discussed throughout this review and are listed below. Not every one of these data types will be available for every drug, gene, variant or phenotype on PharmGKB.
Pathways—Illustrated diagrams of the pharmacokinetics or pharmacodynamics of a PGx‐relevant drug.
VIP summaries—Written reviews of PGx‐relevant genes.
Dosing guidelines—Guidelines from outside organizations on how to adjust treatment of certain medications based on PGx.
Rx annotations—Summaries of individual publications that provide medication dosing or prescribing information based on PGx.
Drug labels—Medication labels containing PGx information.
Variant annotations—Summaries of genetic variant–drug associations as reported in a single publication.
Clinical annotations—Evidence‐rated summaries of all the published evidence for a genetic variant–drug association.
In addition to being freely available on the website, VIPs and pathways are also typically published in the journal Pharmacogenetics and Genomics.
2.2. Pharmacodynamics
While pharmacokinetics encompasses what the body does to the drug, pharmacodynamics can be described as encompassing what the drug does to the body. In the context of PGx, this means the focus is on genes coding for the direct targets of the drug, as well as any gene‐products affected downstream or responsible for the clinical outcome (Karczewski, Daneshjou, & Altman, 2012).
An example of how a genetic variant can affect how the drug works in the body and lead to an adverse reaction can be found by looking at one of the most well‐studied PGx associations: abacavir and HLA‐B*57:01. Abacavir is used for treatment of HIV, and is metabolized into an active form, carbovir 5′‐triphosphate (CBV‐TP), by various enzymes. CBV‐TP blocks the action of HIV reverse transcriptase, which plays a vital role in the replication cycle of the HIV virus (Barbarino, Kroetz, Altman, & Klein, 2014). Abacavir is generally well‐tolerated, however, approximately 5–8% of patents experience a hypersensitivity reaction within the first 6 weeks of treatment, with symptoms such as fever, rash, and gastrointestinal symptoms. These symptoms typically improve within 24 hr of discontinuation, but drug re‐challenge after discontinuation can lead to a potentially life‐threatening allergic reaction. This hypersensitivity reaction is strongly linked with the presence of the HLA‐B*57:01 variant (Barbarino et al., 2014). The HLA‐B protein is responsible for presenting peptides to immune system cells, triggering an immune reaction if the peptide is recognized as coming from a pathogen. Recent evidence suggests that abacavir may bind to the specific form of HLA‐B encoded by HLA‐B*57:01, and when bound, alters the peptides that the protein can present. The altered peptides are perceived as foreign by the immune system, triggering a reaction that presents as the hypersensitivity reaction described above (Illing et al., 2012; Norcross et al., 2012; Ostrov et al., 2012). However, research is still ongoing into the exact mechanism of the abacavir‐triggered hypersensitivity reaction.
Due to the strong linkage between the variant and drug response, testing for HLA‐B*57:01 prior to abacavir treatment has become one of the best examples of pharmacogenetic research being used in the clinic. Both the CPIC (Martin et al., 2014) and the DPWG (Swen et al., 2011) have published guidelines recommending that an alternative agent to abacavir be used in patients who are positive for the HLA‐B*57:01 variant. Additionally, the FDA‐approved drug label for abacavir states that genetic testing for the variant is required prior to initiating or reinitiating treatment with abacavir in patients of unknown HLA‐B*57:01 status, and that it is contraindicated in HLA‐B*57:01‐positive patients (ViiV Healthcare Company, 2015). Abacavir drug labels from the European Medicines Agency (EMA, ViiV Healthcare UK Limited, 2017) and Health Canada (HCSC, ViiV Healthcare ULC, 2015) use language of similar strength—both state that testing for the variant should be done prior to treatment due to the high risk of experiencing a hypersensitivity reaction.
PharmGKB provides annotated versions of all three of these labels on the website (Figure 3). The FDA label was annotated from the list of drug labels containing PGx information maintained by the FDA known as the Table of Pharmacogenomic Biomarkers in Drug Labelling (https://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm). PharmGKB regularly references this list and provides annotations of these labels to users, as well as a version of the original drug label available for download with all relevant PGx content highlighted. Occasionally, PharmGKB curators come across drug labels with PGx content that are not on the FDA's list; these too are added to the website. Each drug label is also given a level of evidence based on the wording regarding genetic testing prior to treatment (more information can be found at https://www.pharmgkb.org/page/drugLabelLegend).
Figure 3.
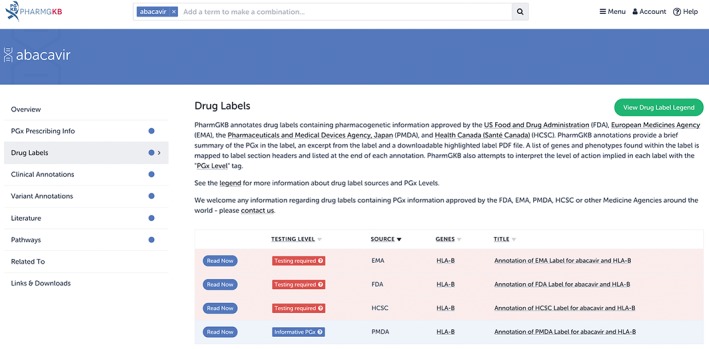
An overview of the annotated drug labels for abacavir on the Pharmacogenomics Knowledgebase (PharmGKB). PharmGKB annotates drug labels approved by the US Food and Drug Administration (FDA), the European Medicines Agency (EMA), Health Canada (HCSC), and the Pharmaceutical and Medical Devices Agency (PMDA), Japan. PharmGKB has annotated drug labels available for over 250 drugs; not every drug has annotated labels available from all four of these medicine agencies
PharmGKB annotates international labels from the EMA and HCSC that correspond to annotated labels from the FDA, if these international labels also contain PGx content. PharmGKB also provides annotated labels from the Pharmaceutical and Medical Devices Agency (PMDA), Japan, which were added with the assistance and translation work of the Japanese Society of Pharmacogenomics (http://www.jspgx.org). Figure 4 provides a visualization of small section of PharmGKB's table of drug labels. By sourcing these international drug labels, PharmGKB provides an informative contrast in how different countries provide PGx information within their labels.
Figure 4.
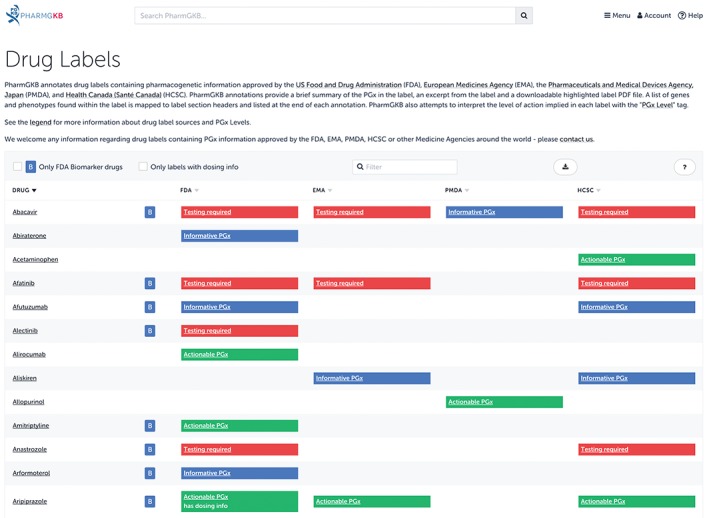
A small section of the drug labels available on the Pharmacogenomics Knowledgebase (PharmGKB). PharmGKB tags Food and Drug Administration (FDA) labels with a B inside a blue square if the label came from the FDA's Biomarker list (https://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm). A small set of labels contain specific dosing guidance based on genetic information; these labels are tagged as including dosing information. For example, in the above image, aripiprazole is tagged in this manner because the FDA label states that CYP2D6 poor metabolizers should have their dose reduced by 50%
Indeed, in the case of abacavir, as seen in Figures 3 and 4, the drug label in Japan contrasts greatly with the Western labels. It provides no guidance regarding testing, stating that the association between the variant and hypersensitivity is unknown in the Japanese population. The label suggests that the absence of an association is due to the low prevalence of the variant in the Japanese—*57:01 which is estimated to have a frequency of only 0.1% in the Japanese as compared to 5–8% in Caucasians. PharmGKB assigned this label as containing “Informative PGx,” indicating that the label mentions a gene involved in the pharmacodynamics, but does not provide any information to suggest that it leads to a different drug response. This contrasts with the “Testing required” given to the FDA, EMA and HCSC labels, which indicates that testing should be conducted before using the drug.
2.3. Somatic variation
While the examples above focused on germline genetic variation—genetic changes that are present in virtually every cell and passed on to offspring—somatic variation is also important within the PGx field. Somatic mutations are those which arise spontaneously within cells (excluding germ cells), and are only passed on to descendants of the original cells that developed the mutation (Griffiths, Miller, Suzuki, et al., 2000). Somatic mutations are involved in the development of cancer—most cancers carry thousands of somatic point mutations and several to hundreds of somatic insertions, deletions, and rearrangements (Martincorena & Campbell, 2015). Anticancer agents can be targeted at this type of cancer‐tissue‐specific genetic variation. For example, the drug vemurafenib (Zelboraf, Genentech, South San Francisco, CA) is indicated for patients with melanoma who have a V600E mutation (1799T>A; rs113488022) within the BRAF protein. This mutation leads to a constitutively activated BRAF protein, resulting in cell proliferation; vemurafenib inhibits this mutated form of the protein. Indeed, the drug is specifically not indicated for patients with wild‐type BRAF, as administration in these patients can instead lead to tumor promotion (Genentech USA, Inc, 2016). A segment of the annotated version of this drug label by PharmGKB can be seen in Figure 5.
Figure 5.
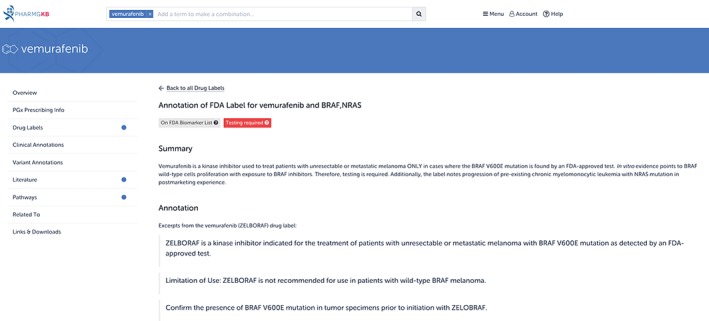
The Food and Drug Administration (FDA)‐approved drug label for vemurafenib (Zelboraf), as annotated on the Pharmacogenomics Knowledgebase (PharmGKB). Vemurafenib is indicated for the treatment of patients with melanoma who have specific somatic mutations. In addition to annotating drug labels containing germline PGx information, PharmGKB also annotates drug labels containing somatic PGx information
Other examples include gefitinib (Iressa, AstraZeneca, Wilmington, DE), which is indicated for non‐small cell lung cancer patients with certain mutations in the EGFR protein (AstraZeneca Pharmaceuticals LP, 2015), and trastuzumab (Herceptin, Genentech, South San Francisco, CA), which is indicated for breast cancer and gastric cancer patients with HER2 protein overexpression (Genentech, Inc, 2015). These two drugs also have annotated labels available on PharmGKB. PharmGKB presents information about cancer pharmacogenomics through the Cancer PGx portal (Figure 6, https://www.pharmgkb.org/page/cancerPgx). PharmGKB primarily focuses on annotating germline PGx associations, including germline genetic variation affecting cancer therapies (e.g., thiopurines and TPMT, discussed earlier in this review), but some information regarding drugs targeted to somatic mutations is available as well. Users can read background on the field, as well as look through all PharmGKB's cancer PGx content.
Figure 6.
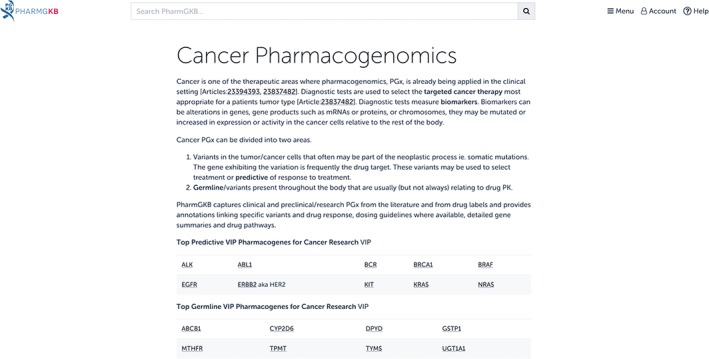
The Pharmacogenomics Knowledgebase (PharmGKB) Cancer PGx portal. Content regarding anticancer drug treatment affected by germline and somatic genetic variation is compiled on a single page for easy browsing
3. PHARMGKB AS A RESOURCE FOR CLINICIANS
While many users of PharmGKB have no exceptional knowledge of PGx or medicine, clinicians make up a significant portion of PharmGKB's user base. Despite the increasing relevance of PGx, multiple barriers currently hinder a greater expansion into the clinic. These include financial considerations, physician education and encouragement, health system infrastructure that can handle genetic data, and the translation of genetic information into actionable recommendations; numerous reviews have detailed these barriers (Dunnenberger et al., 2015). It is important to note that PGx is unlikely to be clinically useful for most prescribed drugs—Relling and Evans show in their 2015 review that only approximately 7% of FDA‐approved medications are affected by “actionable” inherited pharmacogenes, defined by Relling and Evans as those which have the necessary evidence to support analytic validity, clinical validity, and clinical utility (Relling & Evans, 2015). However, they also show that these medications constitute 18% of all prescriptions in the United States, indicating a slight overrepresentation of drugs with actionable PGx information (Relling & Evans, 2015); with the steady increase of published PGx literature, the number of drugs with actionable genetic information is likely to grow.
CPIC aims to address the lack of guidelines for translating genetic information into actionable recommendations. Established in 2009 as a joint effort between PharmGKB and the PGRN, CPIC publishes detailed drug dosing guidelines for clinicians who have access to preemptive genetic test results. The guidelines are freely available, peer‐reviewed and updatable, and include supplementary tables that facilitate translation into machine‐readable electronic health record (EHR) content; more information on CPIC can be found in the 2011 review by Relling and Klein (2011). Other groups, such as the DPWG, and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) also provide gene–drug dosing recommendations; more information on these two organizations can be found in reviews by Swen et al. (2008) and Ross et al. (2010), respectively.
As part of its role in clinical implementation, PharmGKB annotates guidelines by CPIC, DPWG, CPNDS, and other professional groups. However, the knowledgebase also works in collaboration with CPIC to develop its drug dosing guidelines, and provides additional resources to accompany the guidelines on the website. CPIC guidelines are also available through the CPIC website (https://cpicpgx.org/)), where the latest information and most up‐to‐date content for each guideline can be found, as well as comprehensive information on the consortium. CPIC guidelines are written in a standardized format, consisting of background information on the gene, variant and drug being discussed, information on genetic test options, and a discussion of the evidence linking genotype to phenotype. The guidelines culminate in specific therapeutic recommendations written in text form and presented as a table. PharmGKB provides excerpts from the written portion of the CPIC guideline, the full dosing recommendation table and attachments for all guideline and supplementary documents. PharmGKB also enhances CPIC guidelines on its website by providing an interactive version of the dosing guidance; this functionality is shown in Figure 7 for the guideline for TPMT and azathioprine. On any CPIC guideline page, users can enter a genotype or haplotype of interest and receive functional information and dosing recommendations as sourced from the guideline. This tool is useful for guidelines with large or extensive recommendation tables, or users looking for quick dosing information.
Figure 7.
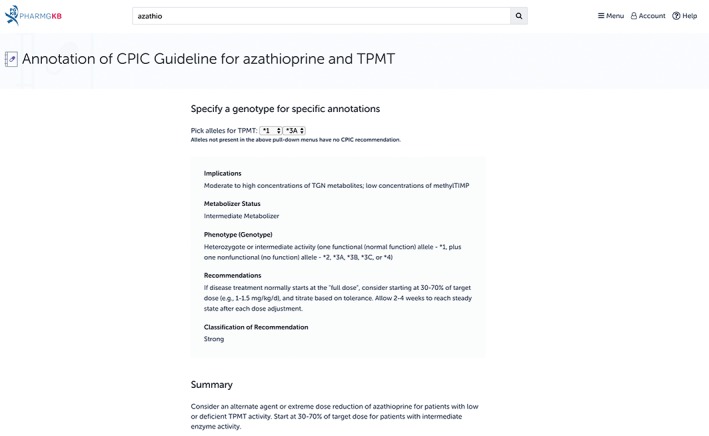
The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for azathioprine and TPMT on the Pharmacogenomics Knowledgebase (PharmGKB) website. In addition to providing the basic content of the CPIC guidelines, PharmGKB also provides its own functionality, allowing users to enter a genotype of interest and receive dosing advice as sourced from the guideline. In the figure, this functionality is shown in action: the TPMT genotype *1/*3A was selected, and detailed information on the implications and dosing recommendations was provided
CPIC guidelines are periodically updated (Caudle et al., 2014); however, occasionally, new information becomes available in between updates that may affect the dosing recommendation. In these instances, CPIC works with PharmGKB to provide a note to users on the guideline page until an updated version is published. PharmGKB also provides gene information tables that support CPIC guidelines, and include spreadsheets detailing star (*) allele mapping, allele functionality information, allele frequency information, and diplotype to phenotype mapping.
Organizations such as CPIC, DPWG, and CPNDS are large consortia, and the peer‐reviewed guidelines they release represent a compilation of knowledge based on extensive review and collaboration. However, individual publications from a single author group can also provide dosing recommendations or algorithms, or results on patient outcomes when these recommendations or algorithms are applied. While these publications may not be as consequential as guidelines from large organizations, they represent clinical implementation at a grassroots level, and are therefore important to provide. These types of studies are termed Rx Annotations on the PharmGKB website, and appear below traditional dosing guidelines from CPIC or DPWG (Figure 8).
Figure 8.
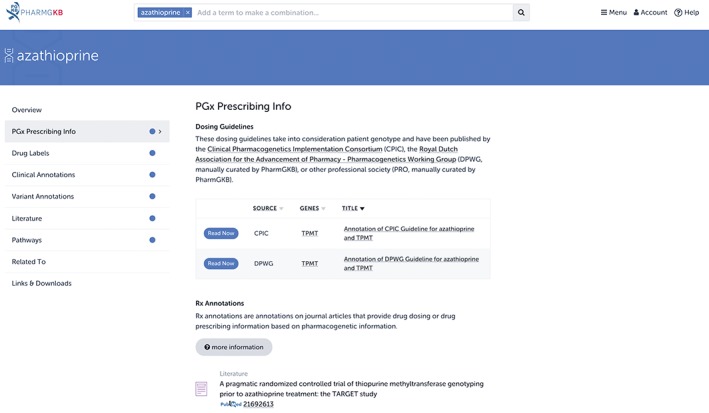
Rx Annotations on the Pharmacogenomics Knowledgebase (PharmGKB) website. In addition to annotating guidelines from organizations such as CPIC and DPWG, PharmGKB also annotates individual publications that provide PGx‐based dosing or prescribing advice. If available, these can be found below dosing guidelines under the “PGx Prescribing Info” tab
4. PHARMGKB AS A RESOURCE FOR PHARMACOGENOMIC RESEARCHERS
When the NIH initiated PharmGKB in April 2000, one of the main goals of the project was to create a repository of primary data, collecting and storing genotype and phenotype data from PGx studies in a systematic manner (Thorn, Klein, & Altman, 2010). The main users of this knowledgebase were scientists interested in PGx associations or data on specific genetic variants (Klein et al., 2001). However, over time, PharmGKB found that the majority of users were more interested in summarized PGx information rather than small primary datasets, and the direction of the knowledgebase shifted from collection of primary data to curation and annotation of peer‐reviewed literature with gene–drug associations. While the mission of PharmGKB has evolved and expanded over time, and the general public and clinicians are now more frequent users, the knowledgebase continues to maintain its collection of annotated literature. Indeed, much of PharmGKB's original content is built upon this collection: drugs or genes that show up repeatedly in the primary PGx literature may warrant a pathway or VIP summary, and PGx associations of strong clinical significance may be candidates for a CPIC guideline. Additionally, PGx data from the primary literature is an invaluable resource for researchers in the field.
PharmGKB stores PGx results from primary literature in the form of variant annotations. These are summaries of an association between a single genetic variant and drug response, such as efficacy or toxicity, as reported in a single publication. While the genetic variation is usually a single nucleotide polymorphism (SNP), PharmGKB also reports on associations for haplotypes, repeats, copy number variations (CNVs), and insertions and deletions (indels). Variant annotations are manually added by curators for both positive and negative results. Each variant annotation contains a standardized summary sentence that describes the results, as well as a free text section and study parameters such as the cohort size, association p‐values and cohort ethnicities (McDonagh, Whirl‐Carrillo, Garten, Altman, & Klein, 2011). Figure 9 shows a variant annotation for the association between HLA‐B*57:01 and abacavir as found in a 2012 study by Berka et al. from a group of patients in Alberta, Canada (Berka, Gill, Liacini, O'Bryan, & Khan, 2012).
Figure 9.
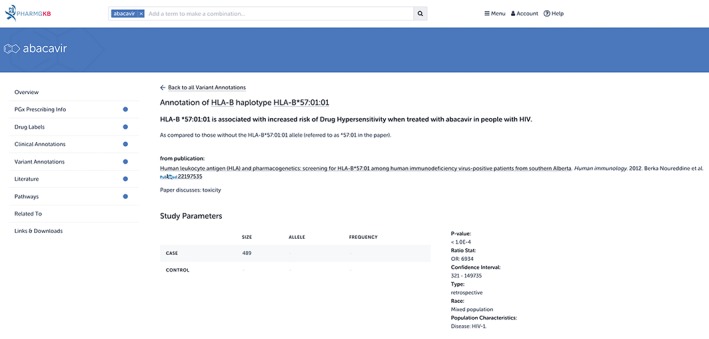
A Pharmacogenomics Knowledgebase (PharmGKB) variant annotation for HLA‐B*57:01 and abacavir. PharmGKB curators read peer‐reviewed literature and add any relevant gene–drug associations to the knowledgebase. Variant annotations consist of a standardized summary sentence describing the association (seen in bold in the figure), as well as descriptive information about the association, such as p‐values, odds ratios, and cohort characteristics
A publication can have as many variant annotations as there are variants and associations discussed. A study may look at associations between multiple genetic variants and the same drug response, in which case each unique variant‐drug association would receive its own annotation. A study may also look at the association between the same genetic variant and different drugs; each variant–unique drug association would receive its own annotation. The enormous number of publications annotated on PharmGKB, combined with the varied number of variant annotations on each publication, leads to a large amount of data.
PharmGKB builds upon variant annotations through the creation of clinical annotations, another unique and valuable resource. Clinical annotations are concise, evidence‐rated summaries of all the literature evidence for a particular genetic variant–drug association. PharmGKB curators manually create clinical annotations by aggregating variant annotations that discuss the same variant and drug phenotype. A short summary is provided that describes the association for each genotype (or haplotype) as compared to other genotypes. Each clinical annotation is assigned a level of evidence, which is based upon factors such as the number of variant annotations showing a positive versus negative results, association p‐values, effect sizes, and cohort sizes. When a new variant annotation is added to a clinical annotation, curators re‐assess the level of evidence, which can increase (or decrease) based on the results. This process allows for the identification of clinically actionable variant–drug associations. Figure 10 shows the clinical annotation for the association between HLA‐B*57:01 and abacavir.
Figure 10.
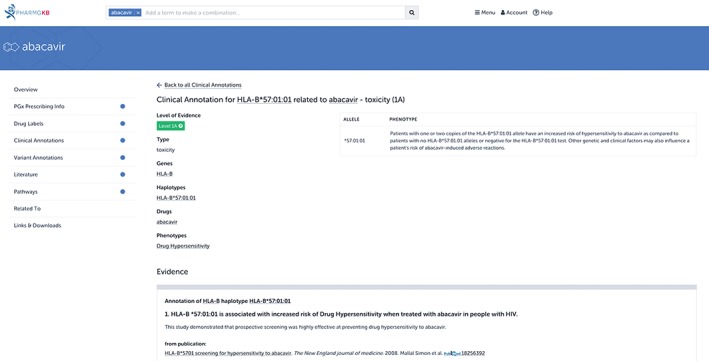
A Pharmacogenomics Knowledgebase (PharmGKB) clinical annotation for HLA‐B*57:01 and abacavir. Clinical annotations accumulate all variant annotations that describe the same gene–drug relationship and present them on a single page with a short summary that describes the association. Each clinical annotation is also given a level of evidence based on the strength of the association. In this example, only one variant annotation is shown as evidence, but there are 12 studies supporting this association: see https://www.pharmgkb.org/clinicalAnnotation/981419257 for the full annotation
All variant and clinical annotations can be easily accessed by casual users on the website. PGx researchers interested in analyzing the data for academic purposes can also access the annotations through bulk download (https://www.pharmgkb.org/downloads) or the Application Program Interface (API) (for more information on technical details of our API, please contact feedback@pharmgkb.org). These downloads are updated monthly. Annotations can be sorted or filtered by variant, gene, drug, or based on the amount of primary PGx data supporting a variant–drug association. Researchers can use the data for such projects as exploring PGx relationships across the genome, predicting new PGx interactions, or providing a PGx perspective on genotype panels or NGS.
4.1. Pharmacogenomics Clinical Annotation Tool (PharmCAT)
While CPIC and other organizations have established guidelines for treatment modifications based on genetic variants, one of the challenges that persist is the ability to easily extract these genetic variants or haplotypes from an individual's whole genome sequence. This is particularly challenging in complex genes such as CYP2D6, which have many named haplotypes composed of several variants. In collaboration with the PGRN Statistical Analysis Resource (P‐STAR), PharmGKB is currently in the process of developing a software tool to assist in this process, known as the Pharmacogenomics Clinical Annotation Tool (PharmCAT, Klein & Ritchie, 2017). The goal of PharmCAT is to extract CPIC guideline variants from a genetic dataset, interpret the variant alleles, and generate a report with actionable dosing recommendations from CPIC. The report can then be used to guide future treatment decisions. PharmGKB is currently in the process of finalizing this tool, with the goal of providing it to the general community for use by the end of the year.
5. CONCLUSION
Despite the growing prominence of both precision medicine and PGx, the universal uptake of PGx in the clinic is still a long way off. While a vast number of articles are published each year detailing associations between genetic variants and drug responses, taking these associations from the primary literature to the bedside of a patient is still a huge challenge. PharmGKB plays an essential role in this process. Its annotation of the primary literature through its variant annotations represents the consolidation of gene–drug data in a format that is easy to aggregate and analyze. Through this aggregation, in the form of its clinical annotations, PharmGKB identifies potentially clinically actionable gene–drug relationships. The organization can then work with CPIC to provide dosing guidelines for these associations. PharmGKB is also anticipating the rise in nonstandardized forms of genetic information that will accompany the decrease in cost of NGS and resultant increase in sequencing by hospitals and other institutions. This includes rare variants that are currently uncatalogued within the published literature, but may be clinically relevant. It remains a future challenge for the knowledgebase to provide support for analysis of these rare variants. However, PharmGKB currently curates diverse types of data in multiple formats, and the knowledgebase expects that this flexibility will enable it to adapt to new forms of genetic information.
While the maintenance and growth of the data contained within the knowledgebase represents a significant portion of PharmGKB's mission, the knowledgebase is also involved in various outside projects that are advancing the field of PGx. This includes PharmCAT, which aims to provide a smoother transition from whole genome results to clinical action, and PharmVAR (http://www.pgrn.org/pharmvar.html), a resource providing standardized nomenclature for genes of particular PGx importance. PGx will transform the way patients are treated for illnesses, reducing the occurrence of dangerous side effects and getting patients the right drug at the right time. PharmGKB will continue to play an integral role in the growth of the field and its further movement into the clinic.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLE
ACKNOWLEDGMENTS
This work was supported by the NIH/NIGMS (R24 GM61374 and R24 GM115264).
Barbarino JM, Whirl‐Carrillo M, Altman RB, Klein TE. PharmGKB: A worldwide resource for pharmacogenomic information. WIREs Syst Biol Med. 2018;10:e1417. https://doi.org/10.1002/wsbm.1417
Funding information NIH/NIGMS, Grant/Award numbers: R24 GM115264, R24 GM61374
References
REFERENCES
- Abbott, A. (2003). With your genes? Take one of these, three times a day. Nature, 425(6960), 760–762. [DOI] [PubMed] [Google Scholar]
- AstraZeneca Pharmaceuticals LP . (2015). IRESSA—gefitinib tablet, coated [package insert]. AstraZeneca Pharmaceuticals LP, Delaware.
- Barbarino, J. M. , Kroetz, D. L. , Altman, R. B. , & Klein, T. E. (2014). PharmGKB summary: Abacavir pathway. Pharmacogenetics and Genomics, 24(5), 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka, N. , Gill, J. M. , Liacini, A. , O'Bryan, T. , & Khan, F. M. (2012). Human leukocyte antigen (HLA) and pharmacogenetics: Screening for HLA‐B*57:01 among human immunodeficiency virus‐positive patients from southern Alberta. Human Immunology, 73(2), 164–167. [DOI] [PubMed] [Google Scholar]
- Caudle, K. E. , Klein, T. E. , Hoffman, J. M. , Muller, D. J. , Whirl‐Carrillo, M. , Gong, L. , … Johnson, S. G. (2014). Incorporation of pharmacogenomics into routine clinical practice: The clinical pharmacogenetics implementation consortium (CPIC) guideline development process. Current Drug Metabolism, 15(2), 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean L. (2012). Mercaptopurine therapy and TPMT genotype In Pratt V, McLeod H, Dean L, et al., eds. Medical genetics summaries. Bethesda, MD: National Center for Biotechnology Information. Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK100660/ [PubMed] [Google Scholar]
- Dunnenberger, H. M. , Crews, K. R. , Hoffman, J. M. , Caudle, K. E. , Broeckel, U. , Howard, S. C. , … Relling, M. V. (2015). Preemptive clinical pharmacogenetics implementation: Current programs in five US medical centers. Annual Review of Pharmacology and Toxicology, 55, 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genentech, Inc . (2015). HERCEPTIN (trastuzumab) for injection, for intravenous use. Genentech, Inc, California.
- Genentech USA, Inc . (2016). ZELBORAF (vemurafenib) [pacakge insert]. Genentech USA, Inc. California.
- Gershon, E. S. , Alliey‐Rodriguez, N. , & Grennan, K. (2014). Ethical and public policy challenges for pharmacogenomics. Dialogues in Clinical Neuroscience, 16(4), 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, D. M. , Cassio, L. , & Livingstone, E. H. (2013). Genomic medicine. Journal of the American Medical Association, 309(14), 1544 Retrieved from http://jamanetwork.com/journals/jama/fullarticle/1677375 23571597 [Google Scholar]
- Griffiths, A. J. F. , Miller, J. H. , Suzuki, D. T. , Lewontin R. C., & Gelbart W. M. (2000). Somatic versus germinal mutation In An introduction to genetic analysis (7th ed.). New York, NY: W.H. Freeman. [Google Scholar]
- Illing, P. T. , Vivian, J. P. , Dudek, N. L. , Kostenko, L. , Chen, Z. , Bharadwaj, M. , … McCluskey, J. (2012). Immune self‐reactivity triggered by drug‐modified HLA‐peptide repertoire. Nature, 486(7404), 554–558. [DOI] [PubMed] [Google Scholar]
- Karczewski, K. J. , Daneshjou, R. , & Altman, R. B. (2012). Pharmacogenomics. PLoS Computational Biology, 8(12), e1002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, T. E. , Chang, J. T. , Cho, M. K. , Easton, K. L. , Fergerson, R. , Hewett, M. , … Altman, R. B. (2001). Integrating genotype and phenotype information: An overview of the PharmGKB project. Pharmacogenetics research network and knowledge base. The Pharmacogenomics Journal, 1(3), 167–170. [DOI] [PubMed] [Google Scholar]
- Klein, T. E., & Ritchie, M. D.. (2017). PharmCAT: A Pharmacogenomics Clinical Annotation Tool. Clinical Pharmacology and Therapeutics. https://www.ncbi.nlm.nih.gov/pubmed/29194583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linköping University, Institutionen för Medicin Och Hälsa . (2013). TPMT alleles. Linkoping, Sweden: Linkoping University. Retrieved from https://www.imh.liu.se/tpmtalleles?l=en [Google Scholar]
- Luzum, J. A. , Pakyz, R. E. , Elsey, A. R. , Haidar, C. E. , Peterson, J. F. , Whirl‐Carrillo, M. , … for the Pharmacogenomics Research Network Translational Pharmacogenetics Program (2017). The pharmacogenomics research network translational pharmacogenetics program: Outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clinical Pharmacology and Therapeutics, 102(3), 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. A. , Hoffman, J. M. , Freimuth, R. R. , Klein, T. E. , Dong, B. J. , Pirmohamed, M. , … Kroetz, D. L. (2014). Clinical pharmacogenetics implementation consortium guidelines for HLA‐B genotype and abacavir dosing: 2014 update. Clinical Pharmacology and Therapeutics, 95(5), 499–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena, I. , & Campbell, P. J. (2015). Somatic mutation in cancer and normal cells. Science, 349(6255), 1483–1489. [DOI] [PubMed] [Google Scholar]
- McDonagh, E. M. , Whirl‐Carrillo, M. , Garten, Y. , Altman, R. B. , & Klein, T. E. (2011). From pharmacogenomic knowledge acquisition to clinical applications: The PharmGKB as a clinical pharmacogenomic biomarker resource. Biomarkers in Medicine, 5(6), 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norcross, M. A. , Luo, S. , Lu, L. , Boyne, M. T. , Gomarteli, M. , Rennels, A. D. , … Buchli, R. (2012). Abacavir induces loading of novel self‐peptides into HLA‐B*57: 01: An autoimmune model for HLA‐associated drug hypersensitivity. AIDS, 26(11), F21–F29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov, D. A. , Grant, B. J. , Pompeu, Y. A. , Sidney, J. , Harndahl, M. , Southwood, S. , … Peters, B. (2012). Drug hypersensitivity caused by alteration of the MHC‐presented self‐peptide repertoire. Proceedings of the National Academy of Sciences of the United States of America, 109(25), 9959–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratain, M. J. , & Plunkett Jr., W. K. (2003). Principles of pharmacokinetics In Kufe D. W., Pollock R. E., Weichselbaum R. R., et al. (Eds.), Holland‐Frei cancer medicine (6th ed.). BC Decker: Hamilton, ON. [Google Scholar]
- Relling, M. V. , & Evans, W. E. (2015). Pharmacogenomics in the clinic. Nature, 526(7573), 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling, M. V. , Gardner, E. E. , Sandborn, W. J. , Schmiegelow, K. , Pui, C. H. , Yee, S. W. , … Clinical Pharmacogenetics Implementation Consortium (2013). Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clinical Pharmacology and Therapeutics, 93(4), 324–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling, M. V. , Gardner, E. E. , Sandborn, W. J. , Schmiegelow, K. , Pui, C. H. , Yee, S. W. , … Klein, T. E. (2011). Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clinical Pharmacology and Therapeutics, 89(3), 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling, M. V. , & Klein, T. E. (2011). CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clinical Pharmacology and Therapeutics, 89(3), 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, C. J. , Visscher, H. , Sistonen, J. , Brunham, L. R. , Pussegoda, K. , Loo, T. T. , … Hayden, M. R. (2010). The Canadian pharmacogenomics network for drug safety: A model for safety pharmacology. Thyroid, 20(7), 681–687. [DOI] [PubMed] [Google Scholar]
- Scott, S. A. (2011). Personalizing medicine with clinical pharmacogenetics. Genetics in Medicine, 13(12), 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swen, J. J. , Nijenhuis, M. , de Boer, A. , Grandia, L. , Maitland‐van der Zee, A. H. , Mulder, H. , … Guchelaar, H. J. (2011). Pharmacogenetics: From bench to byte—an update of guidelines. Clinical Pharmacology and Therapeutics, 89(5), 662–673. [DOI] [PubMed] [Google Scholar]
- Swen, J. J. , Wilting, I. , de Goede, A. L. , Grandia, L. , Mulder, H. , Touw, D. J. , … Deneer, V. H. (2008). Pharmacogenetics: From bench to byte. Clinical Pharmacology and Therapeutics, 83(5), 781–787. [DOI] [PubMed] [Google Scholar]
- The Precision Medicine Initiative . (2015). Retrieved from https://obamawhitehouse.archives.gov/node/333101
- Thorn, C. F. , Klein, T. E. , & Altman, R. B. (2010). Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics, 11(4), 501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ViiV Healthcare Company . (2015). ZIAGEN—abacavir sulfate tablet, film‐coated [package insert]. ViiV Healthcare Company, Canada.
- ViiV Healthcare UK Limited . (2017). Ziagen 300 mg film‐coated tablets [package insert]. ViiV Healthcare UK Limited, UK.
- ViiV Healthcare ULC . (2015) ZIAGEN—abacavir tablets 300 mg (as abacavir sulfate). Abacavir oral solution, 20 mg/ml (as abacavir sulfate) [package insert]. ViiV Healthcare ULC, Quebec.
- Wang, L. , Pelleymounter, L. , Weinshilboum, R. , Johnson, J. A. , Hebert, J. M. , Altman, R. B. , & Klein, T. E. (2010). Very important pharmacogene summary: Thiopurine S‐methyltransferase. Pharmacogenetics and Genomics, 20(6), 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum, R. M. , & Wang, L. (2006). Pharmacogenetics and pharmacogenomics: Development, science, and translation. Annual Review of Genomics and Human Genetics, 7, 223–245. [DOI] [PubMed] [Google Scholar]
- Zaza, G. , Cheok, M. , Krynetskaia, N. , Thorn, C. , Stocco, G. , Hebert, J. M. , … Altman, R. B. (2010). Thiopurine pathway. Pharmacogenetics and Genomics, 20(9), 573–574. [DOI] [PMC free article] [PubMed] [Google Scholar]


