Figure 3.
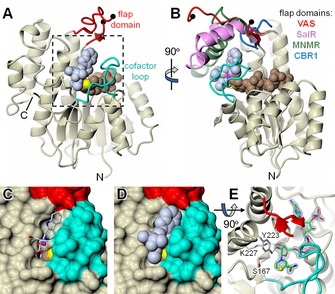
X‐ray structure of C. roseus vitrosamine synthase (VAS). A), B) Orthogonal cartoon representations of VAS, in which the core structure is coloured in magnolia and the flap domain and cofactor loop are shown in red and cyan, respectively. The black spheres indicate where there is a break in the backbone trace of the flap domain corresponding to a region that could not be resolved in the electron density. Also shown as van der Waals spheres are the bound NADP+ cofactor (light brown with C‐4 in yellow) and the docked substrate (ice blue). Additionally, in B), the flap domains of close structural homologues are shown after superposition of their full structures onto that of VAS. Specifically, these are P. somniferum SalR (pink; PDB ID: 3O26), M. piperita MNMR (green; PDB ID: 5L53) and human CBR1 (blue; PDB ID: 1WMA). However, in the last case the flap domain is merely a short loop connecting the equivalent of β4 and α4 in VAS. The full structural superposition is shown as a stereoview in Figure S10. C), D) The region highlighted in A) with the protein depicted as a molecular surface, with and without the docked substrate, respectively. Note that the cofactor loop almost entirely covers the nicotinamide “half” of the NADP+, except for the outer edge of the nicotinamide ring bearing C‐4, and that this is occluded by the docked substrate, which closely matches the dimensions of the active‐site pocket. E) The same region as D), but from above with the protein in cartoon. The NADP+ is coloured green (carbon atoms) but with the nicotinamide C‐4 atom in yellow. Also shown is the canonical SDR catalytic triad of Ser167, Tyr223 and Lys227, together with 1.55 Å resolution omit difference electron density for the cofactor (contoured at ≈5.0 σ). In this view, part of the flap domain was omitted for clarity.
