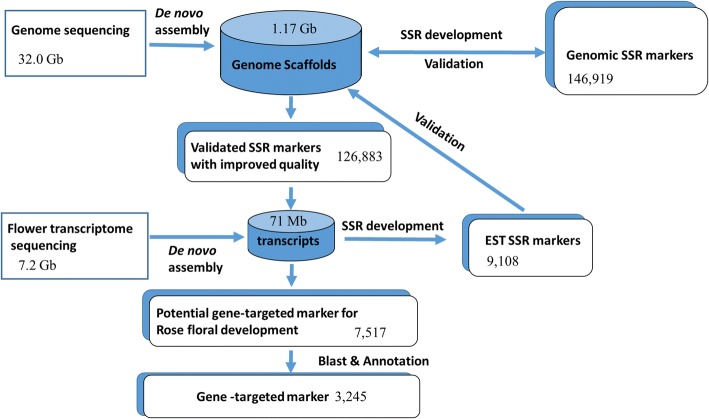
Fig. 6.
Workflow used in the present study. High-throughput sequencing was performed to generate 32 Gb and 7.2 Gb of raw reads for the rose genome and flower transcriptome, respectively. After de novo assembly, a total of 1.17 GB of genomic scaffolds, as well as 71 Mb of EST contigs were obtained. Altogether, 163,680 SSR markers were developed for these genome scaffolds and 16,345 for transcriptomic contigs. Further validation showed that 124,591 genomic and 2762 transcriptomic SSR markers (total of 127,353) had unique annealing sites on the scaffolds; these markers were of better quality than the primary markers. Based on BLAST analysis, 5225 genomic SSR markers were associated with the transcriptome. Together with the EST SSRs, 7987 SSRs were develop that could potentially be used as functional markers for rose floral development