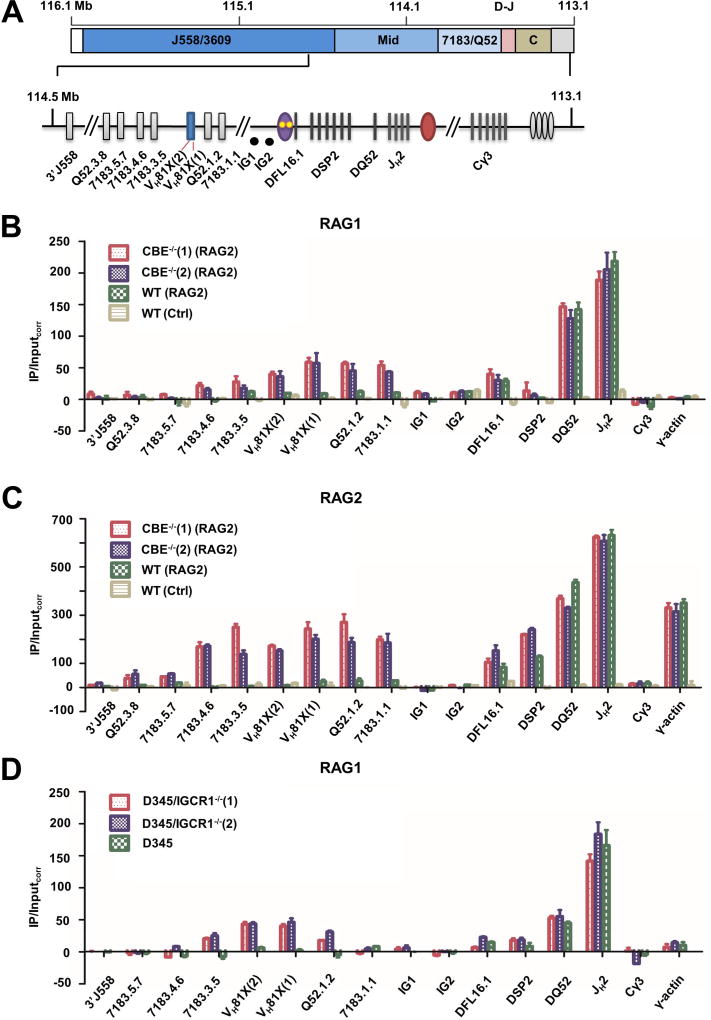
Figure 5. RAG1 and RAG2 recruitment to WT and IGCR1-mutated alleles.
A. Scale schematic of the IgH locus indicating VH gene families (J558/3609, mid, and 7183/Q52), and the 3’ part of the locus containing DH and JH gene segments (D–J), and CH exons (CH). An expanded view of the portion of the locus assayed for RAG1/2 binding by ChIP. ChIP amplicon VH81X (2) scores for unrearranged VH81X only; intergenic amplicons IG1 and IG2 are located approximately 30 kb and 60 kb from VH81X in the 90 kb region between DFL6.1 and VH81X. DSP2 ChIP primers identify 8 DSP2 gene fragments in the unrearranged configuration.
B. RAG1 ChIP. Anti-RAG1 antibody was used to co-precipitate RAG1-associated genomic DNA as described by (Ji et al., 2010) from RAG2-deficient pro-B cell lines that contain IGCR1-mutated (CBE−/−(1) and CBE−/−(2)) or WT IgH alleles (red, purple and green bars, respectively). RAG2 was expressed in these lines by lentiviral transduction. The pro-B cell line with WT IgH alleles was transduced with a puromycin-only lentivirus serving as a control (yellow bars). Co-precipitated genomic DNA and input DNA were assayed by quantitative (real time) PCR and RAG1 enrichment calculated as described by Ji et al. (Y axis). Cγ3 and γ-actin amplicons served as negative controls for RAG1 binding. Two independent RAG2 transductions were carried out, data are shown as mean ± SEM of three independent ChIP experiments analyzed in duplicate for each amplicon.
C. RAG2 ChIP. Anti-RAG2 antibody was used to co-precipitate associated genomic DNA. Cell lines and amplicons are as described in part B. In this case only Cγ3 amplicon served as a negative control, since RAG2 is known to be recruited to the γ-actin promoter in the absence of RAG1. Two independent RAG2 transductions were carried out, data are shown as mean ± SEM of two independent ChIP experiments.
D. RAG1 ChIP in IGCR1-deleted D345 cells. CRISPR/Cas9 was used to delete IGCR1 in D345 pro-B cell line that expresses a catalytically inactive RAG1 and endogenous RAG2. Two cell clones, D345/IGCR1−/−(1) and D345/IGCR1−/−(2), were used for ChIP with anti-RAG1 antibodies. Co-precipitated DNA was analyzed as described in parts B/C with indicated primers. Cγ3 and γ-actin amplicons served as negative controls for RAG1 binding. Data are shown as mean ± SEM of two independent ChIP experiments from each cell line.
See also Figure S7 and Table S3.