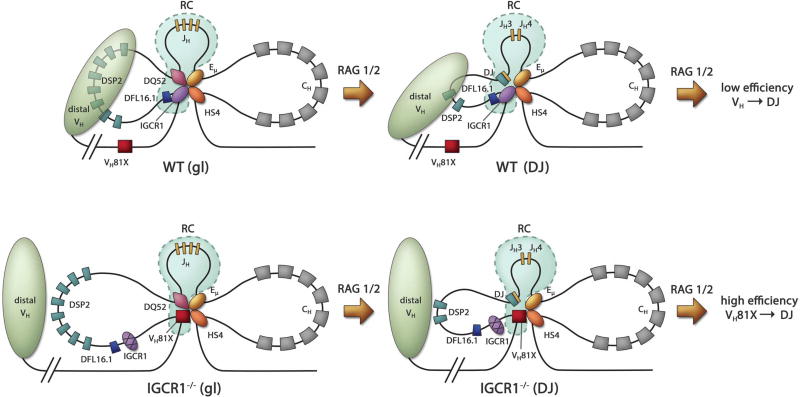
Figure 7. Model for Eμ- and IGCR1-dependent configuration of the 3’ IgH domain.
Top and bottom panels show proposed structures of WT and IGCR1-deficient IgH alleles, respectively, in unrearranged (left) and DJH recombined (right) configurations.
Left: Altered structure and recombination center formation on IGCR1-mutated IgH alleles prior to initiation of recombination (gl = germline alleles). Interactions between IGCR1 (blue oval), the DQ52 promoter (purple diamond), Eμ (yellow oval) and HS4 of the 3’ regulatory region (orange oval) nucleate a 3-loop configuration (top). RAG1/2-rich recombination center (RC) is shown within dotted lines (light blue). The intermediate-sized loop contains all the DH gene segments. The nearest functional VH gene segment, VH81X, is located 90 kb from IGCR1. The compacted domain of distal VH gene segments is shown as a light green oval. Mutation of IGCR1 (bottom) leads to looping of Eμ to the next available CTCF binding site close to VH81X. DFL16.1 lies near the middle of the new 160 kb loop, VH81X is located spatially close to DQ52 and the RC, and the distal VH domain moves away. High levels of RAG1/2 at VH81X reflect an altered RC on IGCR1-mutated alleles.
Right: Proposed structure of WT (top) and IGCR1-deficient (bottom) DJH recombined IgH alleles as discussed in the text.