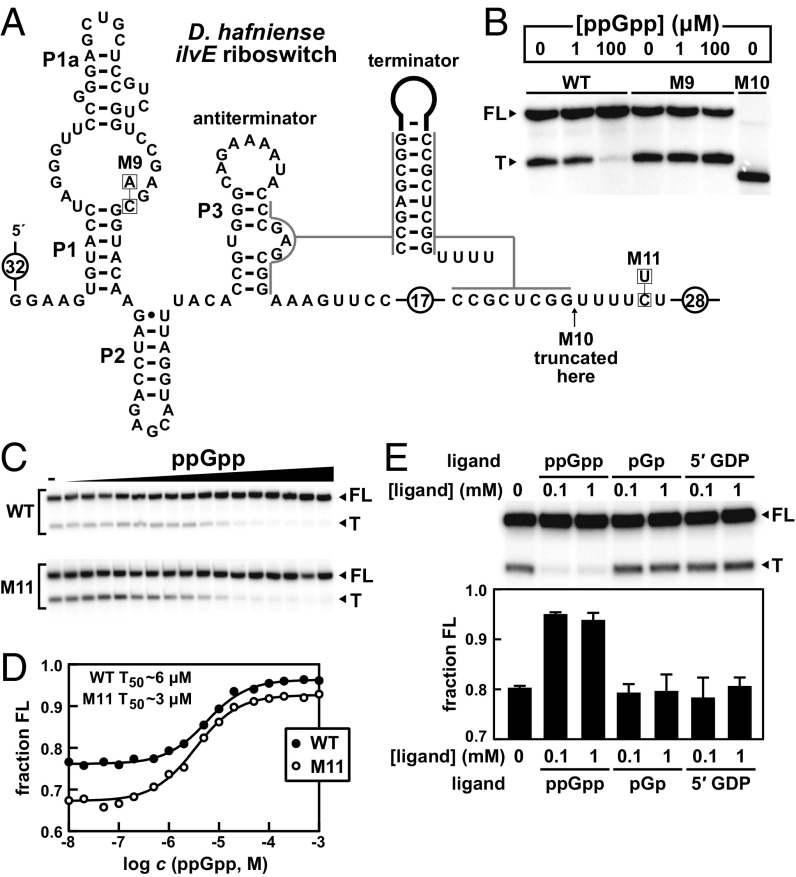
Fig. 4.
ppGpp regulates riboswitch-dependent transcript termination. (A) Sequence and secondary structure of the ppGpp riboswitch derived from the ilvE gene of D. hafniense. An alternative “terminator” RNA structure, which only forms in the absence of ppGpp, is depicted wherein a hairpin forms followed by a U-rich tract. (B) PAGE analysis of single-round transcription termination assays with WT, M9, and M10 D. hafniense ilvE riboswitches conducted in the absence or presence of ppGpp, as indicated. Construct sequences are as depicted in A. FL and T denote full-length and terminated transcripts, respectively. Construct M10 provides a size marker for a transcript that terminates several nucleotides before the product generated when the intrinsic terminator forms. (C) PAGE analysis of single-round transcription termination assays of the WT and M11 D. hafniense ilvE RNAs with either no ligand (–) or ppGpp ranging from 10 nM to 1 mM. (D) Plot of the fraction of FL WT or M11 D. hafniense ilvE riboswitch transcripts contributing to the total number of transcripts (FL plus T) as a function of the logarithm of ppGpp molar concentration. The concentration of ppGpp required to cause half-maximal change in termination (T50) was determined by a sigmoidal curve fit. (E, Upper) PAGE analysis of single-round transcription termination assays of the WT D. hafniense ilvE RNA with either no ligand (–) or each ligand as indicated at a concentration of 0.1 or 1 mM. Data are representative of multiple trials. (Lower) Plot of the fraction of FL WT D. hafniense ilvE riboswitch transcripts contributing to the total number of transcripts (FL plus T) in the absence of ligand (–) or with each ligand as indicated. Values reported are the average and SD of three separate trials, including the PAGE image depicted above.