Significance
Cisplatin is a front-line drug in treatment of most solid tissue cancers. It kills cancer cells by damaging their DNA. Although it is quite effective it has two major drawbacks. First, it has serious side effects, including nephrotoxicity, hepatotoxicity, and neurotoxicity. Secondly, some cancers exhibit primary or acquired resistance to the drug which limit its usefulness. Attempts have been made to administer the drug at certain times of the day (chronochemotherapy) to overcome these limitations but these attempts have had very limited success. Here, we generate genome-wide and at single-nucleotide-resolution circadian DNA repair maps for mouse kidney and liver with the ultimate goal of developing a rational cisplatin chronochemotherapy regimen.
Keywords: cisplatin, chronotherapy, excision repair, mouse, XR-seq
Abstract
Cisplatin is a major cancer chemotherapeutic drug. It kills cancer cells by damaging their DNA, mainly in the form of Pt-d(GpG) diadducts. However, it also has serious side effects, including nephrotoxicity and hepatotoxicity that limit its usefulness. Chronotherapy is taking circadian time into account during therapy to improve the therapeutic index, by improving efficacy and/or limiting toxicity. To this end, we tested the impact of clock time on excision repair of cisplatin-induced DNA damage at single-nucleotide resolution across the genome in mouse kidney and liver. We found that genome repair is controlled by two circadian programs. Repair of the transcribed strand (TS) of active, circadian-controlled genes is dictated by each gene’s phase of transcription, which falls across the circadian cycle with prominent peaks at dawn and dusk. In contrast, repair of the nontranscribed strand (NTS) of all genes, repair of intergenic DNA, and global repair overall peaks at Zeitgeber time ZT08, as basal repair capacity, which is controlled by the circadian clock, peaks at this circadian time. Consequently, the TS and NTS of many genes are repaired out of phase. As most cancers are thought to have defective circadian rhythms, these results suggest that future research on timed dosage of cisplatin could potentially reduce damage to healthy tissue and improve its therapeutic index.
Cisplatin and its derivatives are frontline drugs in the treatment of testicular, ovarian, colorectal, lung, and head and neck cancers (1, 2). However, its utility is compromised by primary or acquired resistance of the cancer cells to the drug and by serious side effects that include nephrotoxicity, hepatotoxicity, and neurotoxicity (3–5). Two approaches have been taken to overcome these limitations. In one approach new derivatives that exhibit lower toxicity or circumvent the DNA repair system that causes resistance have been developed (2, 4). The second approach is chronochemotherapy (6–8). The goal of chronochemotherapy is to deliver the drug at a time of the day that is optimal for killing cancer cells while sparing normal tissue and avoiding development of resistance. For achieving this goal, it is necessary to understand the pharmacodynamics and pharmacokinetic properties of the drug, in particular the factors that impinge upon Pt-DNA adduct formation and repair.
In the past, several attempts have been made to develop chronochemotherapy regimens for combination chemotherapy that included cisplatin or oxaliplatin (6, 7). While some beneficial effects were reported with some of these regimens, the gains have been either minor or have not been replicated in subsequent studies (9). As a consequence, cisplatin chronochemotherapy is currently not practiced in clinical oncology. We reasoned that a mechanism-based approach that takes into account the kinetics of Pt-DNA adduct repair at every single point in the genome including the genes involved in carcinogenic growth and drug resistance may help design a more efficient chronotherapy regimen.
Recently, we developed the eXcision Repair-sequencing (XR-seq) method for whole-genome single-nucleotide-resolution mapping of nucleotide excision repair (10–14). Using this method, we have analyzed the damage and repair maps for UV-, cisplatin-, and oxaliplatin-induced DNA damage in human cells (11, 12). Here, for the first time, we apply XR-seq to generate a repair map from a whole animal using mice as a model system. Moreover, by administering cisplatin and performing XR-seq on kidney and liver at 4-h intervals for a 24-h period, we have generated whole-genome circadian repair maps for these two organs.
Our data reveal that the repair of cisplatin damage in the mouse genome is controlled by two circadian programs. Repair of the transcribed strand (TS) of circadian-controlled genes is dictated by each gene’s phase of transcription (15) and falls across the entire circadian cycle. In contrast, repair of the nontranscribed strand (NTS) of transcribed genes, circadian or otherwise, repair of intergenic DNA, and global repair overall peaks at Zeitgeber time (ZT) ∼ 08 because global repair capacity which is controlled by the circadian clock peaks at this circadian time (16, 17). As a result, the TS and NTS of diurnally expressed genes are repaired out of phase. As it has been reported that cancers may have circadian rhythms in phase with the host (18) or out of phase with the host (19) or even lack rhythmicity (19), these findings constitute a foundation for future research for mechanism-based chronochemotherapy.
Results
High-Resolution, Genome-Wide Mapping of Nucleotide Excision Repair in Mice.
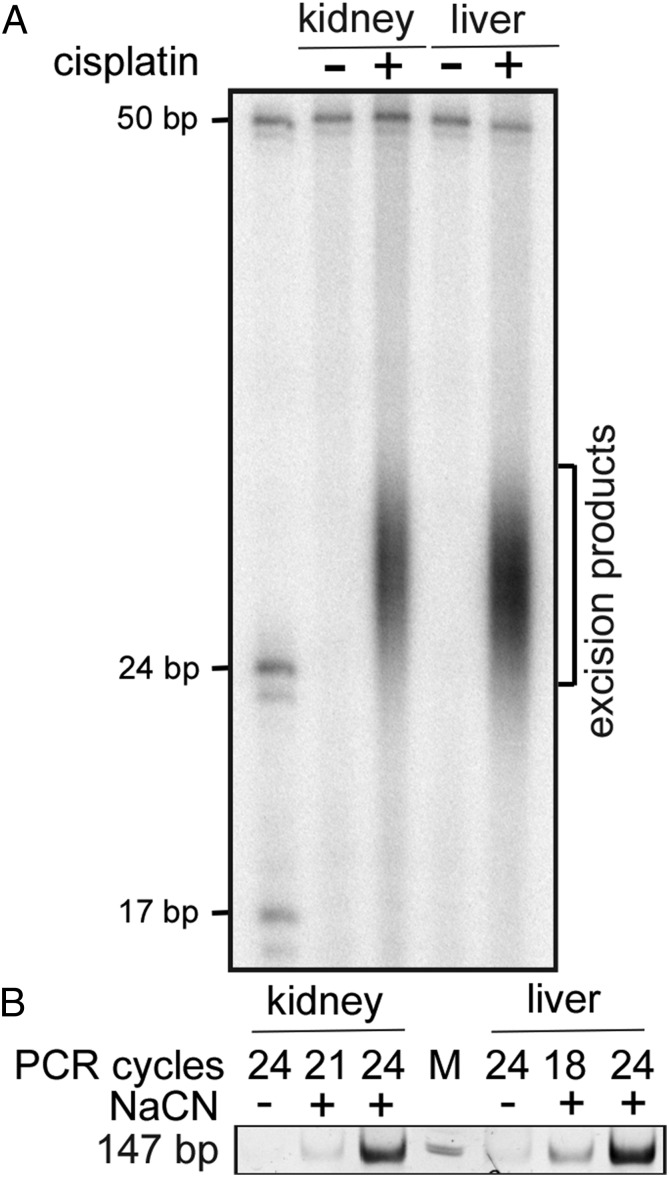
Recently, we developed a nucleotide excision repair (excision repair) assay (XR-seq) for genome-wide mapping of repair sites at single-nucleotide resolution in cultured mammalian cells (11, 12). In this assay, the 26–27-nt-long oligonucleotides generated by the repair reaction are captured, sequenced, and aligned to the genome to generate a repair map. In this investigation, for the first time, we applied this assay to study genome-wide repair in an animal: we injected mice with cisplatin and, after an appropriate period for adduct formation and repair, tissues were harvested and the excision repair products were isolated and sequenced for genome-wide mapping. Fig. 1 shows excision products and XR-seq libraries generated from kidney and liver 2 h after drug administration.
Fig. 1.
Excision products and XR-seq libraries. (A) Detection of the excised oligonucleotides from kidney and liver of mice administered cisplatin at 10 mg/kg. Mice were killed 2 h after cisplatin injection at ZT02. Kidney and liver nuclei were isolated, lysed gently, and TFIIH was immunoprecipitated with anti-p89 and anti-p62 antibodies, which bring down the primary excision products tightly bound to TFIIH (11, 12). The excision products were further purified with anti–Pt-DNA (cisplatin) antibodies. A small fraction of the purified DNA was 3′ labeled with 32P-cordycepin and analyzed on a sequencing gel, along with 17- and 24-nt-long size markers. The 50-mer was included in the labeling reactions as an internal control to measure labeling and recovery efficiency. (B) Analysis of dsDNA libraries generated from the excised oligomers at ZT04. The purified excised oligomers were treated with NaCN where indicated to reverse the Pt adduct and then subjected to the indicated PCR cycles and separated on a polyacrylamide gel (12).
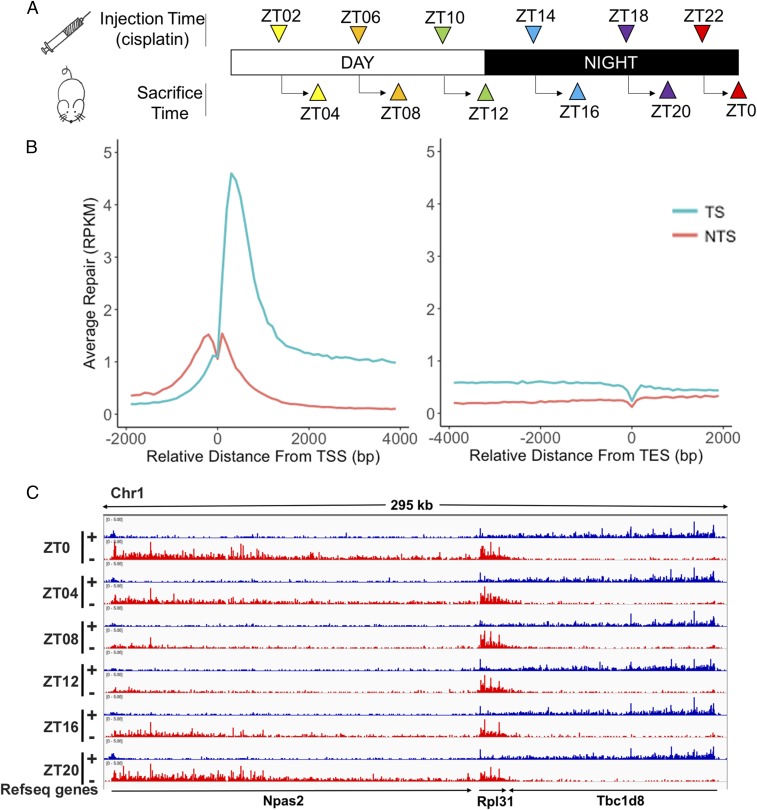
Next, we extended this experiment to generate repair maps as a function of circadian time for mouse kidney and liver. We used the experimental design shown in Fig. 2A. Mice were injected with cisplatin at 4-h intervals, and, 2 h after each injection, kidneys and livers were harvested and excision repair products were isolated and processed by the XR-seq procedure. SI Appendix, Fig. S1 shows that at all circadian time points the excised oligomers are of 26–27-nt median length with the Pt-d(GpG) diadduct located at 19–20/20–21 nucleotides from the 5′ end and 5–6 nucleotides from the 3′ end of the oligonucleotide, in agreement with data from human cells in tissue culture (12–14). Next, we analyzed the effects of genomic location and circadian time on excision repair.
Fig. 2.
Transcriptional and circadian control of excision repair of cisplatin-DNA adduct in mice. (A) Schematic of circadian repair experiment. Mice kept under 12-h light:12-h dark (LD 12:12) condition were administered cisplatin at the indicated time points and tissues were harvested 2 h later; the excision products were isolated and analyzed by XR-seq. ZT indicates circadian time where ZT0 is light-on and ZT12 is light-off. For each time point three mice were killed for XR-seq. Repair data were collected from two independent circadian experiments that were in good agreement (SI Appendix, Fig. S2). (B) Genome-wide analysis of TS and NTS repair shows strong preference for TS repair in promoter-proximal regions, throughout gene bodies, and into the TES. Preferential repair reversal upstream of the TSS is due to bidirectional promoters for most mammalian genes such that the NTS in the gene body becomes the TS upstream of the transcription start sites (TSS). The y axis shows RPKM for 100-nt windows. (C) Illustration showing the combined effects of the circadian clock and transcription on cisplatin repair. Transcription and repair patterns of a 295-kb region of chromosome 1 encompassing the Npas2 clock gene and two neighboring genes are shown. Blue, plus strand XR-seq repair reads; red, minus strand XR-seq repair reads. The Npas2 gene is itself clock-regulated and repair of its TS peaks at ZT20-ZT0 and troughs at ZT08. The clock output gene Rpl31 exhibits much weaker rhythmicity in repair that is delayed compared with Npas2 (peak ZT0-ZT08, minimum ZT16). Tbc1d8 exhibits high amplitude and constant TS repair over the entire course of the circadian cycle.
Overall Effects of Transcription on Repair.
When we analyzed the repair of the TS and NTS genome-wide, we observed results similar to those obtained with cultured cells (12). First, as reported for excision repair in tissue culture (12, 15), the TS is repaired 5–10-fold more efficiently than the NTS (Fig. 2B). Second, as in tissue culture, the promoter-proximal region is repaired with higher efficiency than the gene body because of the high fraction of aborted transcripts in the first 1,000 bp of RNA polymerase II-transcribed genes (20, 21). Third, at transcription end sites (TES) there is a strong dip in repair at the termination site but the TS preference still continues beyond the TES, as for most genes the TES is a region rather than a single point. Finally, upstream of promoters, the strand preference for repair is reversed. This is because for ∼80% of genes the promoter region contains a unidirectional forward core promoter for transcribing the gene and a unidirectional reverse core promoter (21) that is responsible for divergent short upstream transcripts.
Effect of Circadian Clock on Transcription-Coupled Repair.
Previously, using in vitro assays with cell-free extracts, we reported that a key core excision repair factor, XPA, and therefore the cellular repair activity, was controlled by the circadian clock with a peak at ZT ∼ 10 and a trough at ZT ∼ 22 (16). Since the in vitro assay measures global repair and not transcription-coupled repair (13, 15), it was of interest to analyze the contribution of this global repair oscillation on repair of TS and NTS in genes that are controlled by the circadian clock (22) and in genes that are constitutively transcribed. Fig. 2C shows the repair patterns of the TS and NTS of the circadian clock gene Npas2, a nearby clock-controlled gene which is transcribed codirectionally with Npas2, Rpl31 [as reported by MetaCycle (23)], and a constitutively transcribed gene that is transcribed in the opposite direction, Tbc1d8. Repair of the TS of Npas2 is overwhelmingly clock-controlled with a peak at ZT20-ZT0 and a trough at ZT08-12, in agreement with its known transcriptional rhythmicity (22). The NTS of Npas2 is repaired poorly at all circadian times relative to the peak value of the TS. The TS of Rpl31, on the same strand as Npas2, is also highly efficiently repaired relative to its NTS but oscillation of its TS repair is relatively weak and delayed compared with Npas2 (maximum ZT0-ZT08, minimum ZT16), in agreement with its transcriptional profile in MetaCycle. The Tbc1d8 gene, which is expressed constitutively and transcribed in the opposite direction of Npas2, exhibits a high ratio of TS/NTS repair over the entire circadian cycle. Similar results are seen throughout the genome with respect to the repair of the TS and NTS of constitutively expressed and clock-controlled genes over the entire circadian cycle (discussed further below).
Genome-Wide Analysis of Circadian Control of TS and NTS in the Mouse Kidney.
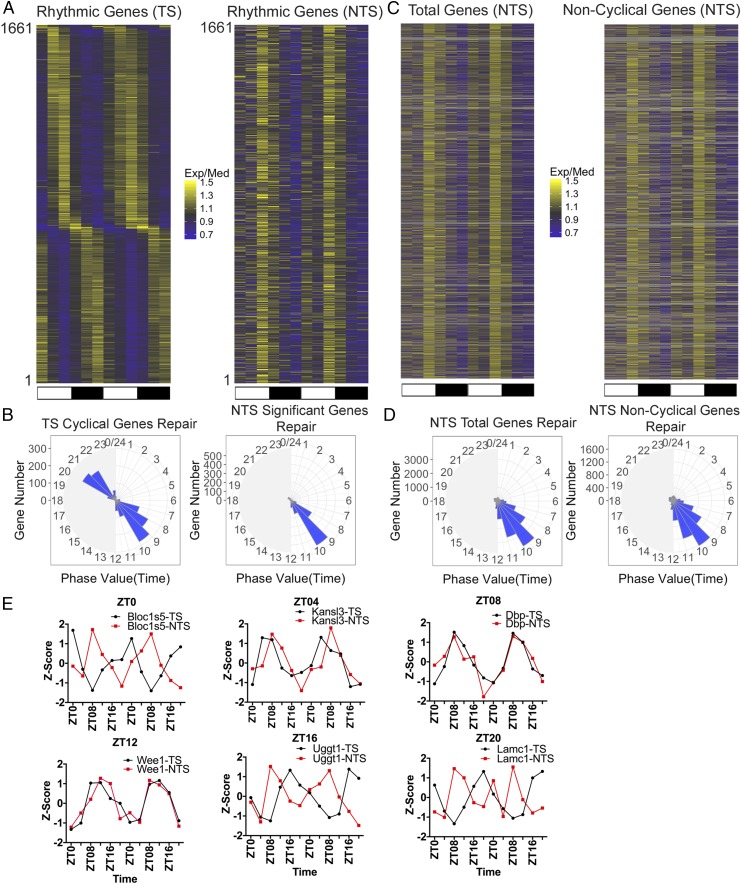
To determine the effect of the circadian clock on TS and NTS repair genome-wide, we analyzed the repair patterns of 1,661 genes that exhibited rhythmicity in TS repair with an amplitude of twofold or higher in kidney. The heatmaps in Fig. 3A reveal different patterns of rhythmicity for the TS and NTS. Repair maxima for the TS are spread over the entire circadian cycle with two major clusters at ZT08 and ZT20. In contrast, NTS repair of the same genes has a single maximum at ZT08. This point is more clearly illustrated by plotting the data in the form of radial diagrams (24) (Fig. 3B). In agreement with several transcription profiling studies (22–32) the TS repair shows two anticipatory circadian peaks at predawn and predusk. The exact phases of these peaks differ slightly from those of the transcriptional analyses, which is most likely due to technical limitations in measuring transcription or repair phase. Indeed, similar differences are seen among the transcriptional profiling reports (25, 28, 31). Of special interest, in contrast to the TS repair, the NTS repair exhibits a single peak at predusk (Fig. 3B), which is consistent with the oscillation of basal excision repair activity with a peak at ZT ∼ 10 (16).
Fig. 3.
Two interdependent circadian programs control repair of the TS and NTS. (A) Heatmaps of circadian repair cycles of TS and NTS of 1,661 highly rhythmic genes in mouse kidney. Exp/Med is, for each gene, RPKM at a given ZT divided by the median ZT RPKM value. Note the distribution of the repair maxima over the entire circadian cycle for the TS and the single maximum for repair of the NTS due to the circadian-controlled peak repair activity which manifests itself on the NTS but its contribution to the TS repair is obscured by the much stronger effect of transcription on repair. The scale for selecting the significant cyclical genes is meta2d_pvalue < 0.05, meta2d_rAMP > 0.1. Each horizontal line represents one gene from ZT0 to ZT24 with two replicates. (B) Radial diagram representation of TS and NTS repair. The TS repair exhibits two peaks corresponding to predawn and predusk, in agreement with numerous transcriptional analyses studies (22, 28, 31). The NTS repair exhibits a single peak at ZT08-11 in agreement with the peak transcription-independent excision repair activity (16, 17). The scale for selecting the significant cyclical genes both in TS and NTS is meta2d_pvalue < 0.05, meta2d_rAMP > 0.1. (C) Heatmap representation of global repair of the NTS of all genes (Left). Genes without significant rhythmicity (based on TS, meta2d_pvalue ≥ 0.05) are shown (Right). Each horizontal line represents one gene from ZT0 to ZT24 with two replicates. (D) Global repair phase of total genes (Left) and noncyclical genes (Right) are presented in radial plots. Analysis of phase distribution reveals peak NTS repair at ZT08-11. (E) Examples of dissonance of the TS vs. NTS repair for genes were chosen to represent transcription (and hence TS repair) maxima covering an entire circadian cycle. The dissonance is most apparent when the transcription/repair phase is farthest from ZT08, which represents the total repair activity and hence maximum NTS repair. We used three animals per time point for analysis and performed two biological replicates. The time range is from ZT0 to ZT24. In E (as in A), data for ZT0 to ZT24 replicate one are followed by data for ZT0 to ZT24, replicate two.
Interestingly, when we plot the repair profiles of all annotated genes, we observe a maximum at ZT08 for the NTS (Fig. 3 C and D) and the removal of rhythmically transcribed genes from this analysis does not change the basic patterns of either the heatmap or the radial map, indicating that the repair maximum at ZT08 for the NTS is the consequence of repair oscillation and is independent of the circadian control of target genes transcription.
For clock-controlled genes (24, 25, 27–30), the effect of transcription oscillation on repair of the TS and the effect of repair oscillation on the NTS give rise to an interesting phenomenon: For many of these genes, the two strands are repaired out of phase with one another (Fig. 3E). Here, we plotted the TS repair z-score of representative genes with repair maxima ranging from ZT0 to ZT20 along with the NTS repair z-score of the same genes. It is apparent that at the single gene level the NTS repair is out of phase with the TS repair, except when the TS repair peak is also at ZT08. This phase dissonance between the two strands is most striking when the TS repair maxima are at ZT0 and ZT20.
Analysis of cisplatin repair in liver yielded results similar to kidney, with TS repair peaks anticipating dawn and dusk and a single peak of NTS repair anticipating dusk (SI Appendix, Fig. S3). However, the NTS repair peak in liver was considerably wider (ZT07-11). This is most likely due to a combination of factors, including the relatively slow adduct formation kinetics in liver compared with kidney (33), the larger number of oscillating transcripts in liver, and the effect of feeding on liver metabolism (31, 32). While further research is needed to deconvolute the contributions of these factors to the liver repair pattern, on the whole, the liver repair profile supports the conclusion that the TS and NTS repair are controlled by different programs.
Effect of the Circadian Clock on Cisplatin Repairome in Mouse.
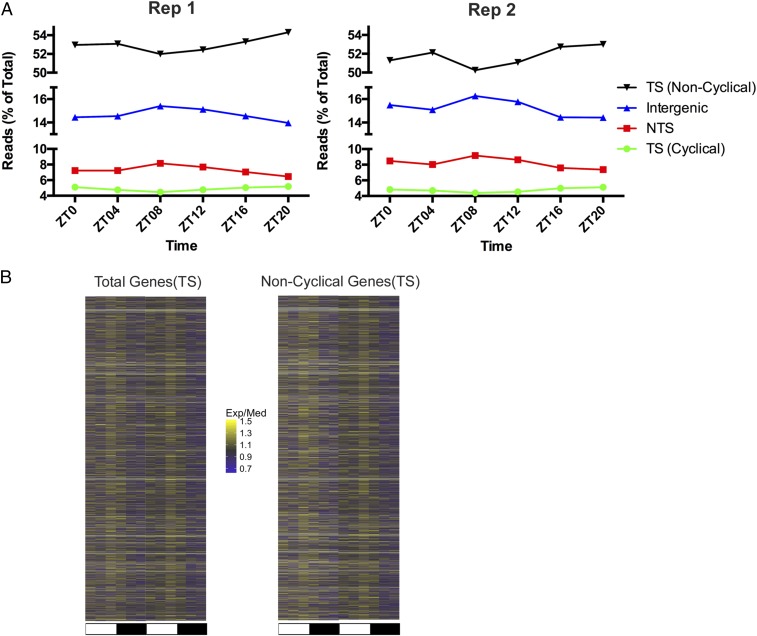
In a previous study, overall excision repair activity in the skin of UV-irradiated mice was measured at two time points, and more repair was observed at ZT09 than ZT21 (17), in agreement with our repair rhythmicity peak at ZT ∼ 08 in the NTS of kidney and liver (Fig. 3 and SI Appendix, Fig. S3). Since the NTS constitutes a minority of the genome, we expected to find this repair program in control of other regions of the genome of our cisplatin-treated mice. In fact, Fig. 4A shows that intergenic regions, which make up a large portion of the genome (SI Appendix, Fig. S4), do show repair rhythmicity similar to the NTS of transcribed genes, peaking at ZT08. Of note, in this plot, since repair in different regions is plotted as a percent of total repair at each time point, and the bulk of repair is rhythmic, there is an apparent trough in repair of the TS in both cyclical and noncyclical genes at ZT08.
Fig. 4.
Genome-wide rhythmicity of repair in kidney. (A) The distribution of repair in TS (noncyclical), TS (cyclical), NTS, and intergenic DNA were plotted to show the reads at each ZT as a percent of total. Rhythmicity in global repair (NTS and intergenic) is indicated by the peak in repair at ZT08. NTS and intergenic do not include overlapping genes. (B) Heatmap representation of TS repair of all genes (Left), and genes without significant rhythmicity (Right) based on TS, meta2d_pvalue ≥ 0.05.
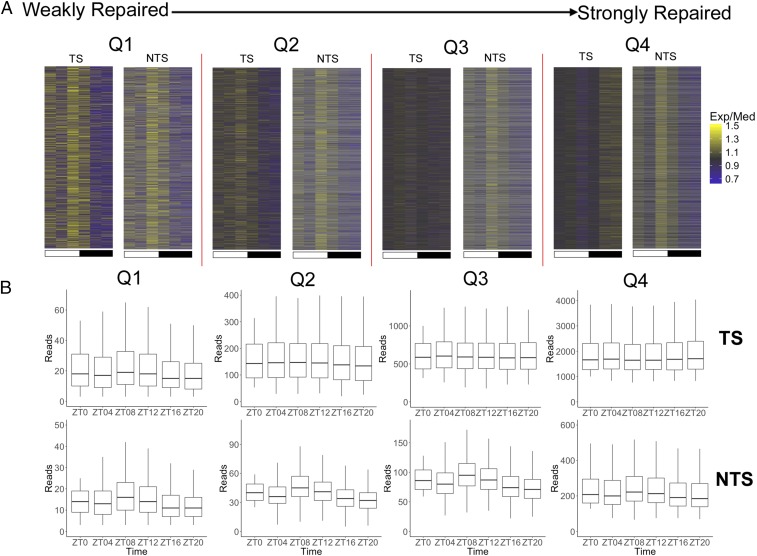
Interestingly, in the plot of TS repair of all annotated genes in Fig. 4B, repair rhythmicity with a peak at ZT08 is seen in the circadian profiles. In this heatmap, repair rhythmicity peaking at ZT08 is observed even with cyclically transcribed/transcription-coupled repaired genes included in the data set, because the latter constitute a minority of all genes, and their peaks at various ZTs tend to obscure one another plotted in this format. Focusing on the noncyclical genes (Fig. 4B, Right), we examined the source of rhythmic TS repair with a ZT08 peak by separating these genes into quartiles based upon repair level (Fig. 5). This analysis shows that TS repair oscillation with a ZT08 maxima is limited to the weakly repaired quartiles (Q1, Q2), which presumably are silent or weakly transcribed. Among the more strongly transcribed genes in Q4, such as Tbc1d8 (Fig. 2C), rhythmicity in repair of the TS mediated by the core repair factors with a peak at ZT08 is not observed because the much more efficient, constitutive transcription-dependent repair predominates. However, for the NTS even in this group the repair peak is at ZT08. The NTS repair peak at ZT08 is less apparent in strongly transcribed genes because it is obscured by the more efficient, constitutive antisense transcription near the promoter. Overall, we find that the rhythmic core repair program with a ZT08 maxima controls large regions of the genome (SI Appendix, Fig. S4) including the TS of weakly or nonexpressed genes, all NTS, and intergenic DNA, and this wide coverage explains the finding that when total repair is measured, a peak at ZT08 is observed (16, 17).
Fig. 5.
Rhythmic repair of the TS of weakly/nontranscribed genes in kidney. (A) Heatmap analysis of repair of noncyclical genes as a function of repair level. After removing cyclical genes, the remaining genes were divided into quartiles Q1 through Q4 having successively higher levels of repair. The TS exhibits repair oscillation in the weakly repaired and presumably weakly- or nontranscribed genes in Q1 and Q2. In contrast, when the repair level and hence transcription is high (Q4), efficient TCR dominates the repair process, and obscures any basal repair activity oscillation; thus, TS repair at ZT08 is at a level comparable to or lower than repair at other time points. In all quartiles repair oscillation is seen in the NTS strand which is as expected from Fig. 3 A and C, and serves as a positive control here. (B) Boxplot representation of the level of total reads of TS and NTS repair in different quartiles, Q1–Q4. For A and B, we limit the minimum reads number plus 2 for each gene at different time points, and each quartile has 3,565 genes.
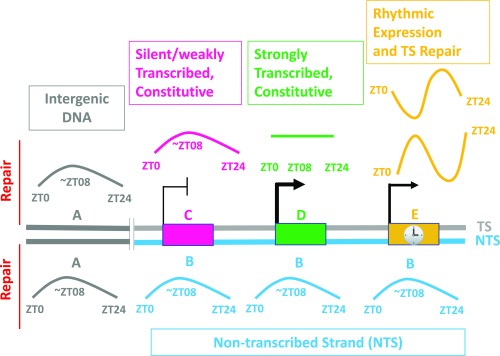
In summary, our findings categorize discrete regions of the genome based upon their circadian program of repair as shown in Fig. 6. In this representation, the rhythmic repair pattern characterized by a peak in repair at ∼ZT08 is due to a peak in activity of the XPA protein and hence basal nucleotide excision repair activity at ∼ZT08 (16, 17). The effect of this program is seen in intergenic DNA, all NTS DNA, and the TS of silent genes and weakly transcribed constitutive genes. Transcription-driven repair is a far more efficient repair mode (13, 15), and genes that are strongly, constitutively expressed are strongly and constitutively (arrhythmically) repaired because transcription-coupled repair (TCR) is much more efficient than the rhythmic repair activity which is mediated by the basal repair factors only. Circadian controlled, rhythmically expressed genes exhibit a striking rhythmic repair of the TS that peaks at different times of day depending upon the expression phase of each gene. The relative abundance of these various categories dictates the repair patterns we have observed for basal genomic repair, for TS and NTS repair of constitutively expressed, and the repair of circadian-clock–controlled rhythmically expressed genes.
Fig. 6.
Model for circadian-clock control of repair of cisplatin damage in the mouse genome. (A) Intergenic DNA; (B) NTS (C) TS-silent/weakly transcribed constitutive genes. (D) TS-strongly transcribed constitutive genes; (E) TS-rhythmically expressed genes. Colored lines indicate levels of repair which may be constant (flat line) or rhythmic (curved and sinusoidal). Rectangles denote genes: the NTS and NTS repair are in blue, the TS is gray, and intergenic DNA and intergenic DNA repair are in dark gray.
Discussion
Here we report the application of the XR-seq method to generate single-nucleotide-resolution DNA repair maps from an animal for the first time. By damaging DNA with cisplatin and sampling repair over an entire circadian cycle, we have obtained circadian repair maps for mouse kidney and liver. These analyses have enabled us to determine which strand of which gene is repaired at a given time of the day for the entire mouse genome and have revealed that for many genes the TS and NTS are repaired out of phase because repair is controlled by two circadian programs: the molecular circadian clock that controls the transcription and TCR of clock-controlled genes which fall across the entire circadian cycle (22), and the global repair activity which is controlled by the clock (16, 17) and exhibits a maximum at ZT ∼ 08. We believe our data, which predict when each strand of all genes will be repaired, provide the potential for developing mechanism-based chemotherapy with cisplatin.
For over three decades, attempts have been made to apply chronochemotherapy to cisplatin-containing combined chemotherapy regimens with mixed, and, at times, contradictory, results. In particular, an early report claimed that application of chronotherapy to combination cisplatin plus doxorubicin treatment of advanced ovarian cancer nearly quadrupled the 5-y life expectancy (34, 35). However, a follow-up clinical trial with a larger group of patients using the same drug combination and chronotherapy regimen did not detect a difference in median duration of survival of patients on standard-timed vs. circadian-timed chemotherapy (36).
More recently, two large chronochemotherapy trials conducted in the United States and in Europe have failed to show significant benefits of cisplatin-containing combined drug regimens in the chronotherapy arm of the trials. The American study (37), by the Gynecologic Oncology Group (GOG), concluded that there was no significant benefit in terms of response rate of progression-free survival, overall survival, or toxicity profile with circadian-timed vs. standard-timed doxorubicin plus cisplatin treatment in patients with advanced or recurrent endometrial carcinoma. Consequently, the GOG has decided not to conduct additional clinical trials of circadian-timed chemotherapy in endometrial cancer.
The European study (38), by the European Organization for Research and Treatment of Cancer Chronotherapy Group examined the combination of Oxaliplatin, Fluorouracil, and Leucovorin as first-line chemotherapy for metastatic colorectal cancer and found that survival and toxicity side effects were similar between patients with circadian-timed and standard-timed treatment. Importantly, the risk of earlier death with chronotherapy vis-à-vis standard therapy was marginally but statistically significantly decreased in men, while the risk of early death in the chronotherapy arm of the women was increased by a larger margin than the small decrease seen in men. Consequently, this chronotherapy regimen is no longer in use. In fact, to our knowledge, there are no chronochemotherapy regimens that have shown beneficial effects and have been subsequently confirmed by other clinical trials.
We believe that instead of empirical approaches, a mechanism-based approach is needed to devise a potentially beneficial cisplatin chronochemotherapy regimen. An approach that takes into account the damage and repair patterns of individual genes over the course of the day both in normal tissue and in cancer is likely to reveal whether cisplatin chronotherapy can improve the therapeutic index. Here, we have documented the circadian repair pattern of cisplatin damage genome-wide and at single-nucleotide resolution in normal mouse tissues. Similar high-resolution circadian repair maps of individual cancer types are needed to determine optimal timing and treatment plans to preferentially kill cancer cells while sparing normal tissues. Currently, there are few studies on the circadian clock of cancers. The findings have ranged from cancer tissue being arrhythmic, to being rhythmic but out of phase with normal tissue, to being rhythmic and in phase with normal tissue (18, 19, 39–44). It is necessary to determine the circadian phase and amplitude relationships between cancer and normal tissues to develop truly mechanism-based chronotherapy regimens. We believe the circadian DNA repair maps we have described in this study are a step in that direction. Currently, we are applying the technology described in this paper to various cancer model systems toward the goal of mechanism-based cisplatin chronochemotherapy. However, we wish to note that in light of the studies reporting a whole spectrum of phase relationship between cancer and normal tissues, it is likely that only a subset of cancer types or cancer patients might benefit from mechanism-based chronotherapy.
Materials and Methods
Mice.
Wild-type animals (6-mo-old C57BL/6J females) were purchased from Jackson Laboratory, and maintained on a light:dark 12:12 schedule for at least 2 wk before sacrifice. ZT0 is the time of light-on and ZT12 is the time of light-off. Mice were handled in accordance with the guidelines of the NIH and the University of North Carolina School of Medicine (Institutional Animal Care and Use Committee).
Cisplatin Injection.
Cisplatin (each milliliter contains 1 mg cisplatin and 9 mg sodium chloride in water for injection; Fresenius Kabi Pharmaceutical Company) was administered by i.p. injection of 10-mg cisplatin/kg.
In Vivo Excision Assay.
Two hours after injecting cisplatin, the mice were killed by carbon dioxide exposure, the liver and kidneys were removed and washed extensively with cold PBS, and then homogenized in 5 mL ice-cold PBS using 15 strokes of a Teflon homogenizer, at which point the plunger moved freely. The homogenized tissues were transferred into 50-mL tubes and pelleted by centrifugation in a centrifuge (Model CL2, cat. no. 004260F; Thermo Fisher) at 2,500 rpm for 4 min, the supernatant was discarded, and the pellets were washed three times to remove fatty material. The pellets were suspended in 5 mL ice-cold Buffer A [25 mM Hepes (pH 7.9)], 100 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12.5% glycerol, 0.5% Nonidet P-40)/per liver or two kidneys and incubated for 10 min on ice. Resuspended cells were transferred to an ice-cold Dounce homogenizer and lysed on ice with 60 strokes using a tight plunger. The chromatin fraction was then pelleted by centrifugation for 30 min at 14,000 rpm at 4 °C in a centrifuge (Model 5418, cat. no. 022620304; Eppendorf). The supernatants were harvested for the first immunoprecipitation [anti-TFIIH, p89 antibody (G-10) and p62 antibody (H-10); Santa Cruz Biotechnology]. The precipitation and wash procedures were described previously (10). The DNAs were subjected to a second round of immunoprecipitation with anti-cisplatin antibody (ab103261; Abcam) (12) before labeling. DNA labeling has been previously described (10) and the labeled DNAs were resolved in 10% denaturing sequencing gels.
XR-Seq Assay.
Treatment of mice, harvesting of tissues, and purification of excision products was the same as in the in vivo excision assay. Excision products were treated with NaCN to remove Pt before PCR (12). Remaining steps, including oligonucleotides and adaptors were according to the previously described XR-seq procedure (11).
Sequencing and Genome Alignment.
At least 8.7 million reads were obtained for each replicate (after removing duplicate reads). Flanking adapter sequences were removed from the reads using BBDuk with command options bbduk.sh ktrim = r k = 21 hdist = 2 minlen = 1 mink = 15. Duplicate reads were removed by FASTX-Toolkit with command options fastx_collapser -v -Q33. Reads were aligned to the mm10 mouse genome using bowtie2 with command options bowtie2 -f–very-sensitive -x -u -s. Following alignment, the files were split into plus and minus strands for subsequent analysis. The raw data and alignment data are available on Gene Expression Omnibus (GEO), accession number GSE109938.
Data Visualization.
For comparing repair signals at different time points, we normalized all of the count data to a sequencing depth of 10 million. The bigwig file is visualized by IGV (software.broadinstitute.org/software/igv/, Broad Institute, and the Regents of the University of California) (45, 46).
Plotting Average Repair Profiles.
To plot average repair profiles relative to annotated transcription start site (TSS) or TES, we used only genes that do not overlap and are at least 5,000 bp in length. Read counts were calculated from aligned bam files using bedtools coverage (47), for 100-nt windows, counts that are normalized to RPKM (reads per kilobase per million).
Analysis of Data from XR-Seq.
Aligned reads were strand-specifically assigned to genes using bedtools with command line options bedtools intersect -c -a -b. The reference gene list mm10 was downloaded from the University of California, Santa Cruz (UCSC) genome browser and was used to remove overlapping genes.
TS gene hit numbers were normalized by RPKM, and then by meta2d (23) with default set except minper = 24, maxper = 24, and cycMethod = c(“JTK,” “LS”). The scale for selecting the significant cyclical genes is meta2d_pvalue < 0.05, meta2d_rAMP > 0.1. To draw the heatmap in Fig. 3A, 1,661 genes from kidney were selected. Relative expression for each gene in Fig. 3A is represented by the RPKM value for each time point divided by the median RPKM value of the six time points. Biological replicates were used and plotted left to right with ZT0 to ZT20 data from experiment 1 followed by ZT0 to ZT20 data from experiment 2. The NTS plot used the same 1,661 genes selected for the TS plot.
For representative genes at different ZT in Fig. 3E, we plot with z-scores calculating by RPKM. z-score is, for each gene, RPKM at a given ZT minus the mean ZT RPKM value divided by the SD of the ZT RPKM value.
Down-Sampled Analysis.
We randomly selected 8.7 million reads (replicate 1) or 20 million reads (replicate 2) from each sample using subsample (https://github.com/paulgb/subsample). The reference genome mm10 was downloaded from the UCSC genome browser. We separated the whole chromatin into gene area, intergenic area, and overlapping gene based upon the reference genes. Then we obtained read numbers for each area. TS and overlapping gene repair are mainly TCR, and NTS and intergenic repair are mainly global repair. After meta2d analysis by using read numbers, we selected significant cyclical genes for making Fig. 4A.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants GM118102 and ES027255 (to A.S.) and NS054794 (to J.B.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw data and alignment data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE109938).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804493115/-/DCSupplemental.
References
- 1.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 3.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 4.Johnstone TC, Suntharalingam K, Lippard SJ. The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs. Chem Rev. 2016;116:3436–3486. doi: 10.1021/acs.chemrev.5b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaney SG, Sancar A. DNA repair: Enzymatic mechanisms and relevance to drug response. J Natl Cancer Inst. 1996;88:1346–1360. doi: 10.1093/jnci/88.19.1346. [DOI] [PubMed] [Google Scholar]
- 6.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 7.Lévi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 8.Paschos GK, Baggs JE, Hogenesch JB, FitzGerald GA. The role of clock genes in pharmacology. Annu Rev Pharmacol Toxicol. 2010;50:187–214. doi: 10.1146/annurev.pharmtox.010909.105621. [DOI] [PubMed] [Google Scholar]
- 9.Sancar A, et al. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, et al. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci USA. 2016;113:11507–11512. doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Selby CP, Adar S, Adebali O, Sancar A. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J Biol Chem. 2017;292:15588–15597. doi: 10.1074/jbc.R117.807453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 16.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci USA. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comas M, Kuropatwinski KK, Wrobel M, Toshkov I, Antoch MP. Daily rhythms are retained both in spontaneously developed sarcomas and in xenografts grown in immunocompromised SCID mice. Chronobiol Int. 2014;31:901–910. doi: 10.3109/07420528.2014.925469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu EA, Weaver DR. Disrupting the circadian clock: Gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2011;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tippens ND, Vihervaara A, Lis JT. Enhancer transcription: What, where, when, and why? Genes Dev. 2018;32:1–3. doi: 10.1101/gad.311605.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo Ngoc L, Wang YL, Kassavetis GA, Kadonaga JT. The punctilious RNA polymerase II core promoter. Genes Dev. 2017;31:1289–1301. doi: 10.1101/gad.303149.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiou YY, et al. Mammalian period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a cryptochrome-dependent manner. Proc Natl Acad Sci USA. 2016;113:E6072–E6079. doi: 10.1073/pnas.1612917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mange F, et al. CycliX Consortium Diurnal regulation of RNA polymerase III transcription is under the control of both the feeding-fasting response and the circadian clock. Genome Res. 2017;27:973–984. doi: 10.1101/gr.217521.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. Time-restricted feeding shifts the skin circadian clock and alters UVB-induced DNA damage. Cell Rep. 2017;20:1061–1072. doi: 10.1016/j.celrep.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dakup PP, et al. The circadian clock regulates cisplatin-induced toxicity and tumor regression in melanoma mouse and human models. Oncotarget. 2018;9:14524–14538. doi: 10.18632/oncotarget.24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrushesky WJ. Circadian timing of cancer chemotherapy. Science. 1985;228:73–75. doi: 10.1126/science.3883493. [DOI] [PubMed] [Google Scholar]
- 35.Hrushesky WJ, Bjarnason GA. Circadian cancer therapy. J Clin Oncol. 1993;11:1403–1417. doi: 10.1200/JCO.1993.11.7.1403. [DOI] [PubMed] [Google Scholar]
- 36.Lévi F, et al. Chemotherapy of advanced ovarian cancer with 4′-O-tetrahydropyranyl doxorubicin and cisplatin: A randomized phase II trial with an evaluation of circadian timing and dose-intensity. J Clin Oncol. 1990;8:705–714. doi: 10.1200/JCO.1990.8.4.705. [DOI] [PubMed] [Google Scholar]
- 37.Gallion HH, et al. Gynecologic Oncology Group Study Randomized phase III trial of standard timed doxorubicin plus cisplatin versus circadian timed doxorubicin plus cisplatin in stage III and IV or recurrent endometrial carcinoma: A Gynecologic Oncology Group study. J Clin Oncol. 2003;21:3808–3813. doi: 10.1200/JCO.2003.10.083. [DOI] [PubMed] [Google Scholar]
- 38.Giacchetti S, et al. European Organisation for Research and Treatment of Cancer Chronotherapy Group Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: The European Organisation for Research and Treatment of Cancer Chronotherapy Group. J Clin Oncol. 2006;24:3562–3569. doi: 10.1200/JCO.2006.06.1440. [DOI] [PubMed] [Google Scholar]
- 39.Sulli G, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature. 2018;553:351–355. doi: 10.1038/nature25170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altman BJ. Cancer clocks out for lunch: Disruption of circadian rhythm and metabolic oscillation in cancer. Front Cell Dev Biol. 2016;4:62. doi: 10.3389/fcell.2016.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiessling S, et al. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017;15:13. doi: 10.1186/s12915-017-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filipski E, et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64:7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 43.Filipski E, et al. Effects of light and food schedules on liver and tumor molecular clocks in mice. J Natl Cancer Inst. 2005;97:507–517. doi: 10.1093/jnci/dji083. [DOI] [PubMed] [Google Scholar]
- 44.Slat EA, et al. Cell-intrinsic, Bmal1-dependent circadian regulation of temozolomide sensitivity in glioblastoma. J Biol Rhythms. 2017;32:121–129. doi: 10.1177/0748730417696788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.