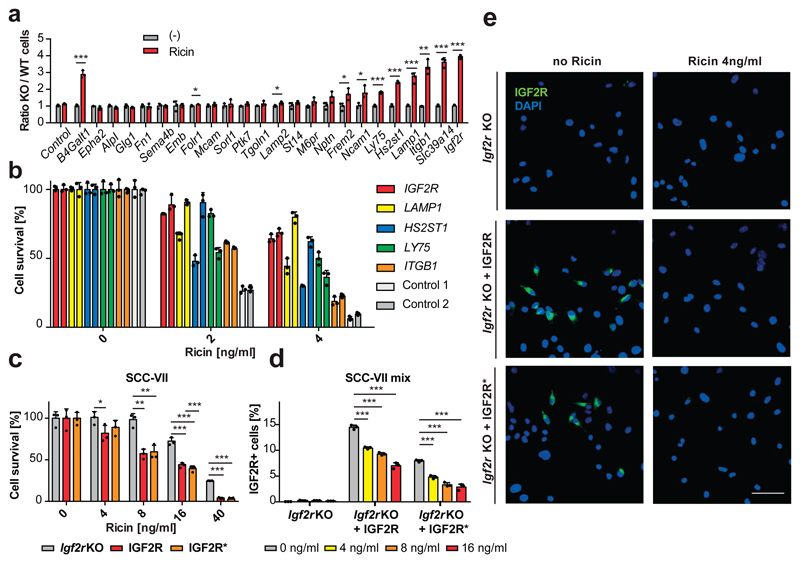
Figure 4. Validation of fucosylated proteins involved in ricin toxicity.
(a) Mixed populations of mutant (KO, GFP+) and their repaired respective wild-type (WT, mCherryCre+) cells were subjected to ricin [4ng/ml] for three days or left untreated (-) and analyzed for viability. The ratios of green to red cells (KO/WT) are shown for each candidate gene. Sister clones for the known ricin sensitivity gene B4Galt131 are shown as positive control and mixtures of GFP and mCherryCre control cells as negative control. (b) HEK cells, harboring CRISPR/Cas9-induced mutations in the indicated genes (2 different sgRNAs each) and control cells (scrambed sgRNAs) were subjected to ricin or left untreated. Cell survival was monitored using Alamar Blue cell viability assay. (c) Igf2r KO murine SCC-VII cells and reconstituted IGF2R wildtype and IGF2R* M6P-binding-domain mutant SCC-VII were subjected to ricin. Cell survival was monitored as above. (d, e) Mixed populations of Igf2r KO and IGF2R/IGF2R* expressing SCC-VII cells were subjected to ricin and the relative amount of IGF2R expressing cells was determined using flow cytometry (d) and confocal microscopy (e). Scale bar 50 μm. Each image is representative of six images of three independent experiments. Data in a, b, c, d are shown as mean ± SD of triplicate cultures. Experiments were repeated three times with similar results. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student’s t-test).