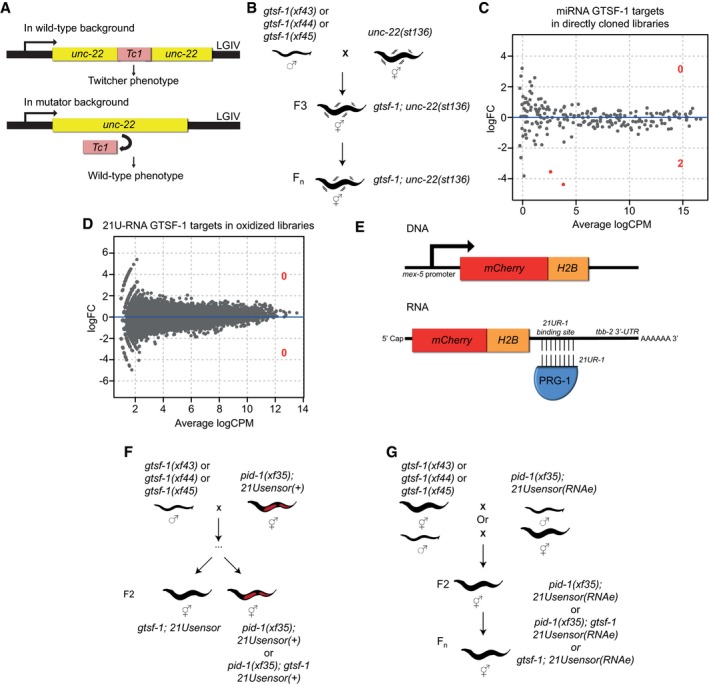
Schematic of the unc‐22(st136) allele.
Layout of the unc‐22(st136) x gtsf‐1 crosses to address transposon derepression. No phenotypic reversions to wild‐type were observed in ten replicate gtsf‐1;unc‐22(st136) populations, grown in parallel for several generations.
Differential gene expression analysis of miRNAs in wild‐type versus gtsf‐1 mutant worms, to address whether miRNAs are globally deregulated in gtsf‐1. Analysis was performed in the directly cloned libraries, given the higher number of miRNA reads observed. Only two miRNAs (mir‐260, mir‐262) are significantly downregulated (1% FDR) in gtsf‐1 mutants.
Differential gene expression analysis of 21U‐RNAs in wild‐type versus gtsf‐1 mutants. Analysis was performed in the oxidized libraries where a larger number of 21U‐RNA reads are found. No significant changes were found at 1% cutoff.
Overview of the 21Usensor. It consists of an mCherry‐histone H2B fusion transgene with a binding site for 21U‐R1, an abundant 21U‐RNA, in the 3′UTR of the transcript.
Testing the participation of gtsf‐1 in 21U‐RNA‐mediated silencing of C. elegans. To test this, a non‐stably silenced 21Usensor was used, and after crossing with gtsf‐1 mutant animals, the F2 of the indicated genotypes was scored for mCherry expression.
Schematics of the crosses between gtsf‐1 mutant alleles and an RNAe 21Usensor. No derepression of the 21Usensor was observed in gtsf‐1 mutants in the F2 and in further generations.