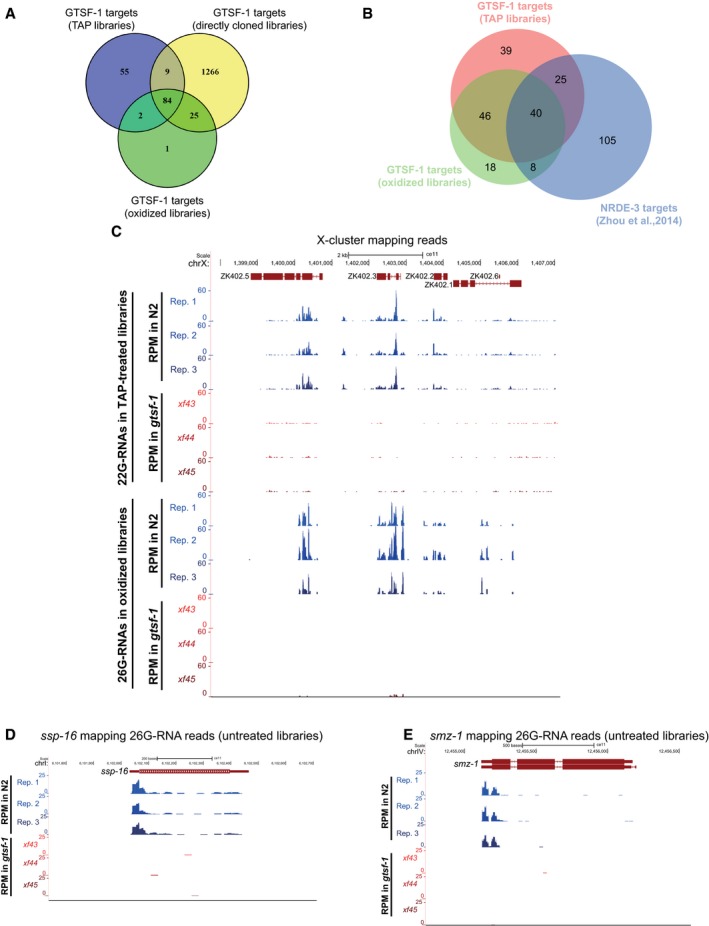
Figure EV4. 26G‐RNAs are severely depleted in gtsf‐1 mutants (related to Fig 4).
-
AVenn diagram showing overlap between the 1% FDR GTSF‐1 targets defined in the three different NGS library preparation conditions.
-
BVenn diagram depicting the overlap between the GTSF‐1 targets defined in the oxidized and TAP‐treated libraries in this study, and NRDE‐3 targets as defined elsewhere (Zhou et al, 2014).
-
C–EExemplary UCSC genome browser tracks with RPM‐normalized sRNA coverage profiles. Three biological replicates of wild‐type N2 worms (Rep. 1–Rep. 3) and the three gtsf‐1 alleles are shown separately. (C) 22G‐RNAs (above) and 26G‐RNAs (below) mapping to the 22G‐RNA X‐cluster. 22G‐ and 26G‐RNA tracks were obtained from TAP‐treated and oxidized libraries, respectively. (D, E) 26G‐RNA reads mapping to ssp‐16 (D), a sperm‐specific family, class P protein, and to smz‐1 (E), a sperm meiosis PDZ domain‐containing protein. Directly cloned libraries were used for these tracks.
