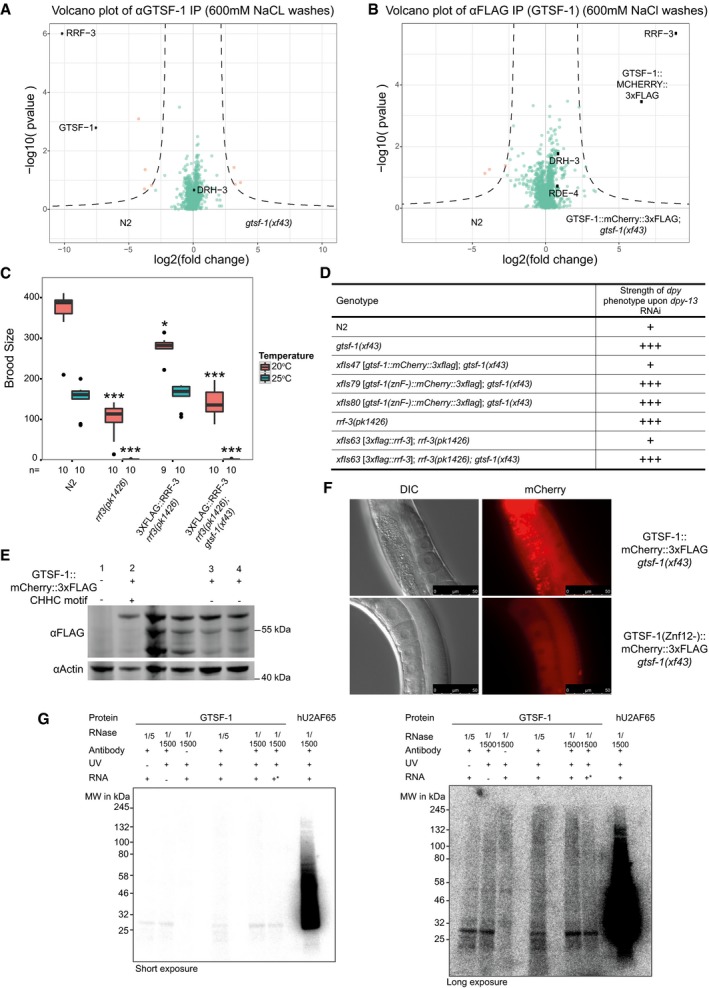
Figure EV5. GTSF‐1 stably interacts with RRF‐3 via its tandem CHHC zinc fingers (related to Figs 5 and 6).
-
A, BVolcano plots of GTSF‐1 IP‐mass spectrometry experiments in more stringent wash conditions (600 mM NaCl). Gravid adult worms were used. IPs were performed and measured in quadruplicates. Other than the wash conditions, the setup was identical to Fig 5A and B.
-
CBrood size count of a 3xFLAG::RRF‐3 single‐copy transgene. This transgene rescues the fertility defects associated with rrf‐3 mutation almost to a complete extent. Asterisk indicates P‐value = 0.002185; triple asterisk indicates P‐value < 0.0001817. P‐values calculated with Mann–Whitney and Wilcoxon tests, using N2 brood size as a reference. n is indicated in the x‐axis. Horizontal lines represent the median, the bottom and top of the box represent the 25th and 75th percentile. Whiskers include data points that are less than 1.5 × IQR away from the 25th and 75th percentile.
-
DOverview of dpy‐13 RNAi experiments to worms of the indicated genotypes. A “+” sign indicates that these animals show a normal response to dpy‐13 RNAi, while “+++” animals have an enhanced response to RNAi, having both a more prevalent and stronger dpy‐13 phenotype.
-
EWestern blot analysis of adult animal populations carrying GTSF‐1::mCherry::3xFLAG transgenes. Lane 1, Wild‐type, non‐transgenic worms. Lane 2, WT GTSF‐1 protein. Lanes 3 and 4 represent GTSF‐1 zinc finger mutants, wherein the zinc finger cysteines are mutated to alanines. The middle, non‐labeled lanes are of other GTSF‐1 fusion proteins not discussed in this work.
-
FWide‐field DIC and fluorescence microscopy pictures of worms expressing WT GTSF‐1::mCherry::3xFLAG (above) and zinc finger mutant GTSF‐1::mCherry::3xFLAG (below). The overall localization of mCherry is not dependent on the zinc fingers. Scale bars indicate 50 μm.
-
GIn vitro iCLIP experiment. Purified GTSF‐1 protein was incubated with Caenorhabditis elegans total RNA from wild‐type, UV cross‐linked and immunoprecipitated using the GTSF‐1 antibody used throughout this study. After the IP, the co‐purified RNA was radioactively labeled and the IPs were run on a SDS–PAGE gel followed by membrane transfer. As a positive control, human U2AF65 (hU2AF65) was used (see Sutandy et al, 2018). GTSF‐1 protein does not associate with RNA beyond background levels. Also, there are not differences between the no antibody, the no cross‐link and the no total RNA controls. Same gel is shown, on the left with short exposure and on the right with longer exposure. +* indicates that gtsf‐1(xf43) total RNA was used, not wild‐type. The rationale behind this was that in the gtsf‐1 mutant background, GTSF‐1 targets are upregulated. This was expected to increase the iCLIP signal.
Source data are available online for this figure.
