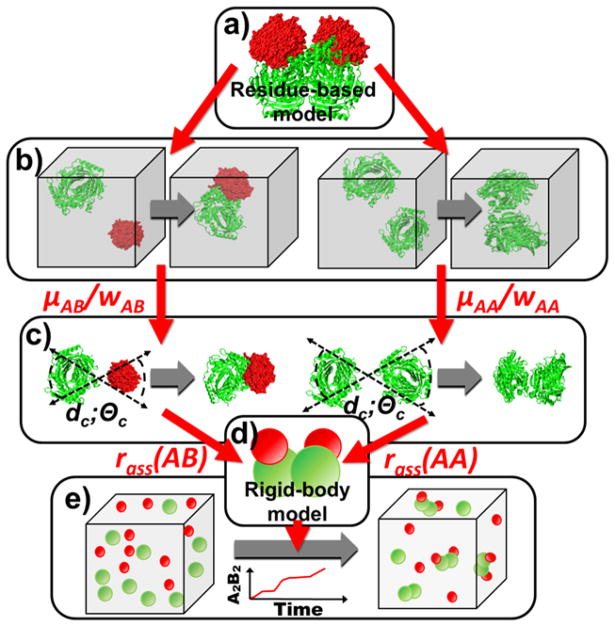
Figure 1.
There are two levels of models in the schematic framework of our multiscale simulation method. The structural details of each protein subunits in a complex can be reflected by the residue-based model (a). We first adjust the parameters in the energy functions of residue-based simulation to reproduce the experimentally measured values of kon for each pair of subunits (b). Given the same energy parameter, the rate of association rass for each pair of subunits is then estimated by the same residue-based model, but with a different boundary condition (c). Finally, the derived values of rass, together with the diffusion constants and binding affinities of interacting subunits, are used to guide the simulation with a rigid-body-based representation (d). The rigid body simulations which contains a large number of protein subunits in the system are able to provide the kinetic information about complex assembly, such as how many final complexes or kinetic intermediate are formed along simulation time (e).