Figure 1. R-loops accumulate in WASp-deficient Th1-cells.
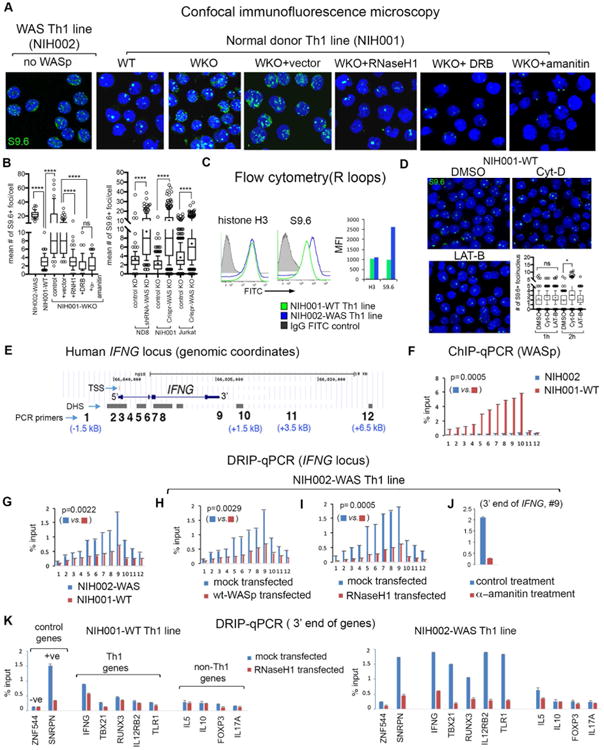
(A) confocal microscopy images of the indicated Th1-skewed cells under indicated experimental conditions dual-labelled with S9.6-antibody (R-loops, green) and DAPI (nucleus, blue). Displayed images are the collapsed composites of 20-30 z-stack images acquired at ×63 magnification. (B) Box-and-whisker plots (whiskers@10-90%, horizontal bar denotes median, “+” denotes mean) generated from 3D (z-stack) images of at least n=100 cells from n=3 independent experiments. (C) Flow cytometry. Histogram plots and bar graphs of their mean fluorescence intensity (MFI) for total histone-H3 (control) and S9.6 (R-loops) in the indicated Th cells. (D) confocal images of S9.6+ foci (R-loops) and their abundance (box-and-whisker plot) in normal donor Th1-skewed cells treated with Cytochalasin-D (Cyt-D), Latrunculin-B (LAT-B), or DMSO-control; n=100 cells from n=3 independent experiments. p-values: ****<0.0001; *=0.01; ns, not significant using Mann-Whitney 2-tailed nonparametric analyses. (E) Genomic locations of 12 PCR primer/probes across the human IFNG-locus (UCSC-Hg18) showing distances (kB) from the 5′TSS and 3′TTS. (F) ChIP-qPCR profile of WASp-enrichment across the IFNG-locus in indicated Th1-cells/conditions. (G-J) DRIP-qPCR profiles of R-loop frequency across the IFNG-locus in Th1-skewed, indicated Th-cells/conditions. (K) DRIP-qPCR: R-loop frequencies at 3′-ends of the indicated genes in the indicated Th1-cells, transfected with RNaseH1 or control vector. Displayed data is the mean of n=3 independent experiments, +SEM; shown are actual Mann-Whitney p-values that compare all data points for the entire IFNG-locus in one condition relative to other. Note: in this study, SNRPN is used as a positive-control for R-loops as previously described (primer coordinates, Chr15:25223714-2522381, in the SNRPN mRNA-sequence, region1-2).32 In another study, an upstream region of the SNRPN-SNURF locus (Chr 15: 25132924-25132943, not in the SNRPN mRNA-sequence) was used as a negative-control for R-loops. 87
