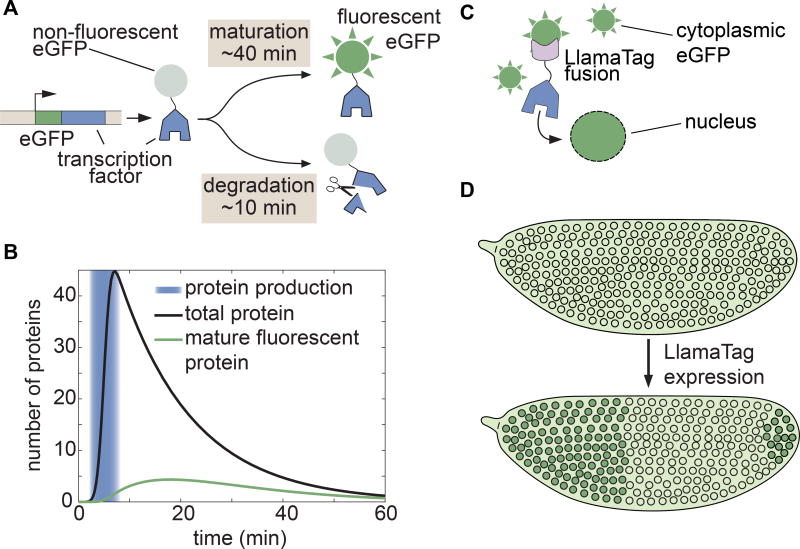
Figure 1. Genetically encoded LlamaTags overcome slow fluorescent protein maturation.
(A) In many model organisms such as the fly, fluorescent protein fusions are degraded much faster than the rate at which they mature and begin to fluoresce. (B) Fluorescent protein maturation masks protein concentration dynamics qualitatively and quantitatively. Total fusion protein includes molecules with both mature and non-mature GFP; the number of mature, visible fluorescent fusion proteins due to a pulse of protein production is plotted over time. Maturation and degradation rates are as in (A); see STAR Methods for model assumptions and parameters. (C, D) In the LlamaTag approach, the fluorescent protein is maternally produced, ensuring that all proteins are fluorescent and uniformly distributed throughout the embryo. Upon translation of a LlamaTagged transcription factor, the fluorescent protein binds the nanobody. Here, eGFP is translocated to the nucleus as a result of the transcription factor’s nuclear localization signal, resulting in the enrichment of nuclear fluorescence. See also Figure S1.