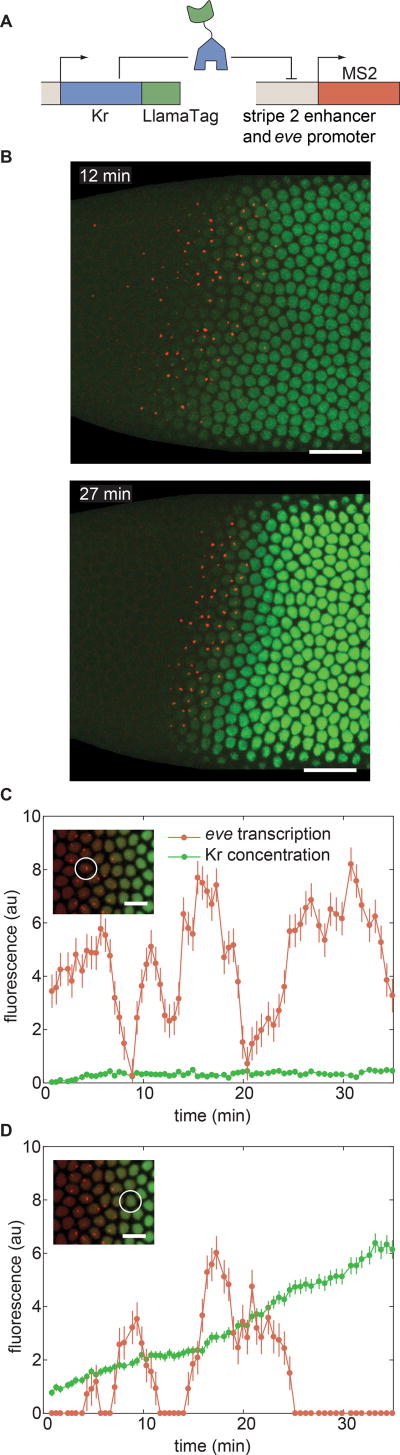
Figure 7. Uncovering the dynamics of transcription-factor input and transcriptional output at the single-cell level.
(A) Strategy for LlamaTagging the Kr repressor and monitoring its regulation of eve stripe 2 transcription using MS2. (B) As time progresses, the increase in concentration of Kr-LlamaTag repressor (green) sharpens the posterior boundary of the stripe 2 of eve transcription (red). Scale bars are 25 µm. The embryo is oriented with anterior to the left and ventral on the bottom. (C,D) Input Kr concentration and output eve transcription for nuclei (C) within the stripe and (D) in the Kr repression domain reveal continuous transcriptional bursts at low Kr concentration, as well as less transcriptional bursting at high Kr concentrations. The scale bars in insets correspond to 10 µm. Error bars for transcriptional activity are estimated from the uncertainty in quantifying the fluorescence background. Error bars for protein concentration are standard error of the intensity values within a nucleus. See STAR Methods for further details. See also Movie S6.