Abstract
Burkholderia pseudomallei is a flagellated, gram-negative environmental bacterium that causes melioidosis, a severe infectious disease of humans and animals in tropical areas. We hypothesised that B. pseudomallei may undergo phenotypic adaptation in response to an increase in growth temperature. We analysed the growth curves of B. pseudomallei strain 153 cultured in Luria–Bertani broth at five different temperatures (25 °C–42 °C) and compared the proteomes of bacteria cultured at 37 °C and 42 °C. B. pseudomallei exhibited the highest growth rate at 37 °C with modest reductions at 30 °C, 40 °C and 42 °C but a more marked delay at 25 °C. Proteome analysis revealed 34 differentially expressed protein spots between bacterial cultures at 42 °C versus 37 °C. These were identified as chaperones (7 spots), metabolic enzymes (12 spots), antioxidants (10 spots), motility proteins (2 spots), structural proteins (2 spots) and hypothetical proteins (1 spot). Of the 22 down-regulated proteins at 42 °C, redundancy in motility and antioxidant proteins was observed. qRT-PCR confirmed decreased expression of fliC and katE. Experiments on three B. pseudomallei strains demonstrated that these had the highest motility, greatest resistance to H2O2 and greatest tolerance to salt stress at 37 °C. Our data suggest that temperature affects B. pseudomallei motility and resistance to stress.
Introduction
Burkholderia pseudomallei is the causative agent of melioidosis, a severe and often fatal disease of humans and animals that is endemic in tropical regions of Asia and northern Australia1. The bacterium can be isolated from soil and water in melioidosis-endemic regions2–4. Infection occurs through skin inoculation, inhalation and ingestion and can be difficult to eradicate1, and B. pseudomallei can remain dormant in humans for a prolonged period1. In northeast Thailand, melioidosis accounts for approximately 20% of all community-acquired septicaemia and is associated with a 40% mortality rate. The disease has a wide spectrum of clinical syndromes but the major manifestations are sepsis, bacteraemia, pneumonia and abscesses in multiple tissues and organs1.
B. pseudomallei has a large genome compared with those of many other species, with two chromosomes of 4.07 Mbp and 3.17 Mbp5. Multiple genes support adaptation and survival in different environments5. B. pseudomallei has been reported to survive in distilled water without nutrients for at least 16 years6. Within the infected host, B. pseudomallei becomes intracellular and can persist in a range of cell types including phagocytic cells, surviving the phagolysosome environment where it is exposed to significant oxidative stress7. B. pseudomallei encodes numerous virulence factors including cell-associated and secretory factors such as type 3 and type 6 secretion systems, proteinases, lipase, lipopolysaccharide, capsule, biofilm and flagella8. Many of these are regulated by bacterial and environmental factors including quorum-sensing systems9,10, VirAG two-component regulatory system11, acid12, salt stress13 and nutritional availability14.
The factors that provide a conducive environment for the survival of B. pseudomallei in adverse conditions are poorly understood. Many bacterial species have altered expression of stress response proteins and repair enzymes as a strategy to reduce temperature-associated damage15–19. This inducible resistance to heat may be essential for bacterial survival in the host. Patients with melioidosis have protracted fever, which is induced by pro-inflammatory mediators produced by the host immune response against bacterial infection. However, it is not known whether high temperature in vivo aids bacterial dissemination, chronic infection or inhibits bacterial multiplication. This indicates a need to understand growth and phenotypic adaptations of B. pseudomallei to heat stress, which requires a discriminatory tool at a proteomic level. We previously used a proteomic approach to characterise and compare protein expression profiles of three different colony morphotypes of B. pseudomallei and described complex changes associated with morphotype switching, which lends support to the idea that this is associated with a fitness advantage under stress environments in vivo12.
We hypothesised that B. pseudomallei undergoes phenotypic adaptation in response to high temperatures in vivo. This study aimed to compare the growth rate of B. pseudomallei cultured at five different temperatures and to use proteomic methods to determine differential protein expression, combined with quantitative reverse transcriptase PCR (qRT-PCR) to confirm protein expression. The effect of different temperatures on phenotypic changes in B. pseudomallei were also examined, including cell motility, flagellar expression, resistance to oxidative and salt stress and biofilm formation. These data provide novel insights into phenotypic changes in B. pseudomallei that may be associated with pathogenesis in vivo.
Results
Effect of temperature on the growth rate of B. pseudomallei
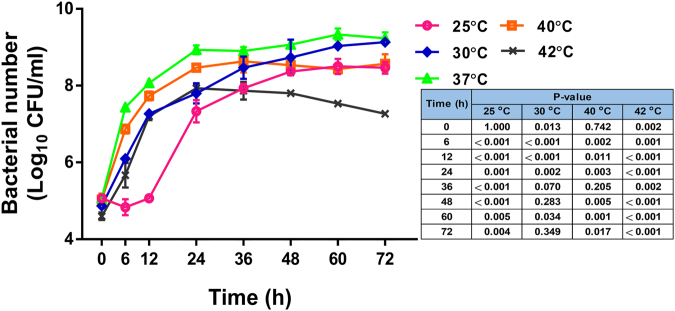
Temperature affects bacterial growth and protein stability20. We investigated the effect of different temperatures on the growth rate of a clinical isolate of B. pseudomallei from a patient with melioidosis, strain 153 in Luria–Bertani (LB) broth at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C. Growth curve analysis revealed that B. pseudomallei exhibited the highest growth rate at 37 °C (Fig. 1). B. pseudomallei grew at 30 °C, 40 °C and 42 °C, albeit at a lower rate. The average doubling times at log phase were 97.7, 90.1, 46.1, 59.5 and 84.0 min for 25 °C, 30 °C, 37 °C, 40 °C and 42 °C, respectively (25 °C versus 37 °C, P = 0.004; 30 °C versus 37 °C, P < 0.001; 37 °C versus 40 °C, P = 0.001; 37 °C versus 42 °C, P = 0.001). The number of viable bacteria cultured at 42 °C gradually fell over time after 24 h of incubation. Exponential growth was delayed until 12 h of incubation for B. pseudomallei cultured at 25 °C, with stationary phase ultimately reached at 48 h. These data indicate that B. pseudomallei can grow and survive in a broad range of temperatures.
Figure 1.
Growth curves of B. pseudomallei strain 153 in LB broth at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C. P values of bacterial number at different temperatures at different time points relative to 37 °C are shown in the table.
Proteomic alteration in B. pseudomallei under heat stress conditions
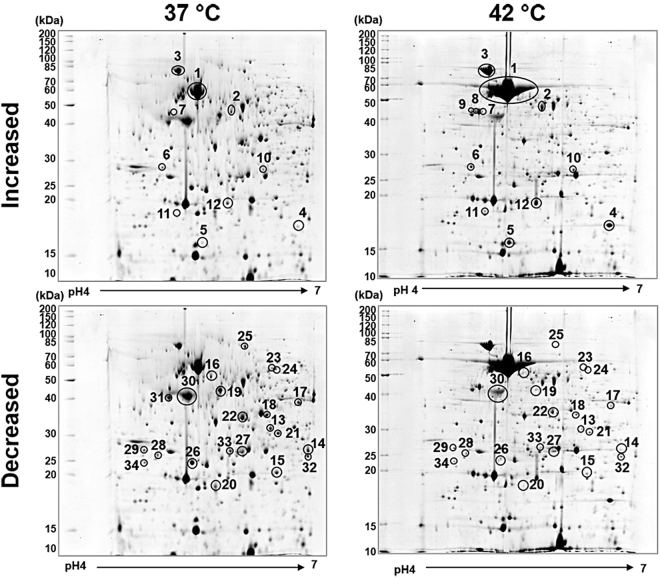
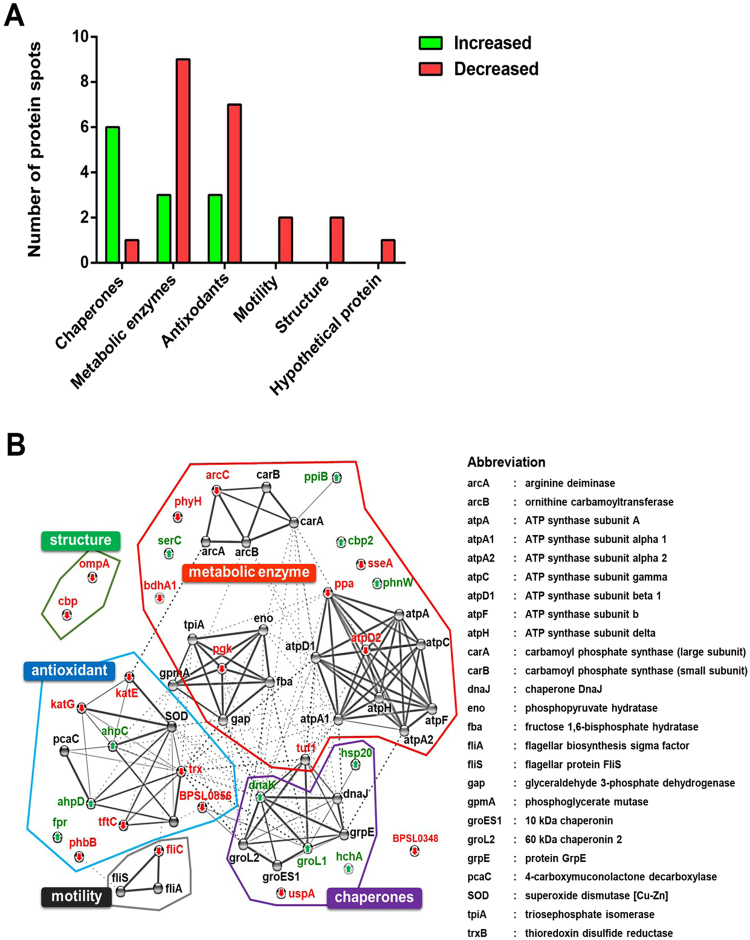
We analysed the proteomic changes of B. pseudomallei strain 153 grown at 37 °C and 42 °C by two-dimensional (2D) gel electrophoresis. Approximately 600 protein spots were visualised after Coomassie blue staining, quantitative intensity analysis of which revealed 34 differentially expressed protein spots with ≥ two-fold change between B. pseudomallei cultured at 42 °C versus 37 °C (Fig. 2 and Table 1). Of these, 12 had increased intensity (2.2 to 24.8 fold) and 22 had decreased intensity (2.3 to 19.0 fold). Two spots were present only at 42 °C, and one spot was absent at this temperature. These proteins were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and were then categorised based on main biological function using UniProt Knowledgebase (Swiss-Prot and TrEMBL entries) (Table 1). Proteins with altered expression were identified as chaperones (7 spots), metabolic enzymes (12 spots), antioxidants (10 spots), motility proteins (2 spots), structural proteins (2 spots) and a hypothetical protein (1 spot).
Figure 2.
Proteomic profiles of B. pseudomallei strain 153. Circles are used to highlight up-regulated proteins (top) and down-regulated proteins (bottom) in bacteria cultured at 42 °C (right) compared with those cultured at 37 °C (left). Protein spot numbers related to information provided in Table 1.
Table 1.
Summary of altered protein expression of B. pseudomallei strain 153 cultured in LB broth at 42 °C compared to 37 °C.
| Protein name | Protein ID | Locus ID | Gene | Biological function | Spot number | MASCOT score | Mass (Da) | PI | Fold change |
|---|---|---|---|---|---|---|---|---|---|
| Upregulated proteins at 42 °C | |||||||||
| Chaperones | |||||||||
| 60 Kda chaperonin | Q9F712 | BPSL2697 | groEL | protein folding/stress response | 1 | 134 | 57137 | 5.13 | 2.97 |
| 2 | 159 | 57137 | 5.13 | 2.73 | |||||
| Chaperone DnaK | O68191 | BPSL2827 | dnaK | protein folding/stress response | 3 | 200 | 69773 | 4.94 | 2.72 |
| Peptidyl-prolyl cis-trans isomerase B | Q63SS5 | BPSL2246 | ppiB | protein folding | 4 | 210 | 17890 | 6.39 | 17.82 |
| Heat shock HSP-20 related protein | Q63QV6 | BPSL2918 | — | stress response | 5 | 89 | 16053 | 5.18 | 24.83 |
| Chaperone protein HchA | A0A0H2WBH2 | BMAA1406 | hchA | stress response | 6 | 213 | 23932 | 4.72 | 2.77 |
| Metabolic enzymes | |||||||||
| Serine-type carboxypeptidase protein | A3N570 | BMA2026 | cbp2 | proteolysis and cellular protein catabolic process | 7 | 56 | 66090 | 5.77 | 3.67 |
| Serine-pyruvate aminotransferase | Q63NF6 | BPSS0343 | phnW | phosphonate and phosphinate metabolism | 8 | 101 | 38917 | 6.30 | Present |
| Phosphoserine aminotransferase | Q63S02 | BPSL2519 | serC | glycine, serine and threonine metabolism | 9 | 134 | 39425 | 5.94 | Present |
| Antioxidants | |||||||||
| Ferredoxin-NADP(H) reductase | Q63YE7 | BPSL0241 | fpr | antioxidant enzyme | 10 | 155 | 28983 | 5.78 | 2.22 |
| Hydroperoxidase reductase | Q63T73 | BPSL2096 | ahpD | antioxidant enzyme | 11 | 87 | 20463 | 5.05 | 2.76 |
| AhpC/Tsa family, antioxidant | Q62I24 | BMA2066 | tsa1 | antioxidant | 12 | 88 | 23904 | 5.75 | 3.63 |
| Down regulated protein at 42 °C | |||||||||
| Chaperone | |||||||||
| Universal stress protein family domain protein | Q2T4Y8 | BTH_II1566 | uspA | stress response | 13 | 129 | 30371 | 5.84 | −5.22 |
| Metabolism enzymes | |||||||||
| Acetoacetyl-CoA reductase | Q63J00 | BPSS1916 | phbB | poly-hydroxybutyrate biosynthetic process | 14 | 190 | 26583 | 6.30 | −4.10 |
| D-beta-hydroxyburyrate dehydrogenase | Q62CL0 | BMAA0017 | bdhA-1 | ketone body biosynthetic process | 15 | 83 | 28137 | 5.90 | −5.78 |
| ATP synthase F1, beta subunit | Q63IW3 | BMA2957 | atpD-2 | ATP hydrolysis coupled proton transport | 16 | 131 | 50819 | 5.26 | −6.41 |
| Elongation factor Tu | Q63PZ6 | BPSL3215 | tuf | protein biosynthesis | 17 | 150 | 43192 | 5.36 | −3.47 |
| Non-ribosomally encoded peptide/ polyketide synthase | Q63L25 | BPSS1183 | phyH | biosynthetic process | 18 | 102 | 35611 | 5.77 | −2.63 |
| Phosphoglycerate kinase | Q63WU5 | BPSL0796 | pgk | carbohydrate metabolic process | 19 | 186 | 41379 | 5.58 | −6.32 |
| Inorganic pyrophosphatase | Q63W67 | BPSL1021 | ppa | phosphate-containing compound metabolic process | 20 | 88 | 19206 | 5.37 | −19.03 |
| Rhodanese-related sulfotransferase | Q63JF0 | BPSS1766 | sseA | thiosulfate sulfurtransferase activity | 21 | 127 | 31119 | 5.98 | −3.82 |
| Carbamate kinase | Q63U71 | BPSL1745 | arcC | arginine metabolic process | 22 | 129 | 33507 | 5.54 | −2.27 |
| Antioxidants | |||||||||
| Catalase HPII | Q63I56 | BPSS2214 | katE | hydrogen peroxide catabolic process/stress response | 23 | 152 | 78455 | 5.89 | −6.80 |
| 24 | 130 | 78455 | 5.89 | −4.19 | |||||
| Catalase-peroxidase proteins | Q939D2 | BPSL2865 | katG | hydrogen peroxide catabolic process/stress response | 25 | 215 | 81824 | 5.89 | −13.67 |
| Oxidoreductase | Q63RC4 | BPSL2748 | tftC | antioxidant enzyme | 26 | 172 | 23904 | 5.75 | −4.44 |
| 27 | 95 | 23904 | 5.75 | −3.22 | |||||
| Thioredoxin | Q63WN5 | BPSL0856 | — | antioxidant enzyme/redox homeostasis | 28 | 233 | 33259 | 4.55 | −3.64 |
| Thioredoxin protein, putative | Q63WN5 | BPSL0856 | trxA | antioxidant enzyme/redox homeostasis | 29 | 62 | 33352 | 4.71 | −4.12 |
| Motility | |||||||||
| Flagellin | H7C7G3 | BPSL3319 | fliC | bacterial flagella | 30 | 106 | 39233 | 5.05 | −2.60 |
| 31 | 120 | 39233 | 5.05 | Absent | |||||
| Structure | |||||||||
| OmpA family protein | Q62M19 | BMA0436 | ompA | structural molecule/ion transmembrane transport | 32 | 109 | 24291 | 9.51 | −2.61 |
| Chitin-binding protein, putative | Q63PN3 | BPSL3340 | cbp | structural molecule | 33 | 110 | 25944 | 6.23 | −2.94 |
| Miscellaneous | |||||||||
| Hypothetical protein | Q63Y40 | BPSL0348 | — | — | 34 | 60 | 28041 | 6.59 | −2.39 |
The 22 proteins with decreased expression at 42 °C were metabolic enzymes (9 spots), antioxidants (7 spots), bacterial motility (2 spots), structural proteins (2 spots), a chaperone (1 spot) and a hypothetical protein (1 spot) (Fig. 3A). By contrast, the 12 proteins with increased expression at 42 °C were mostly related to heat shock protein families and chaperones (6 spots), antioxidants (3 spots) and metabolic enzymes (3 spots) (Fig. 3A). Analysis of global protein networks using STRING revealed associations between these proteins produced at different levels (Fig. 3B), which were consistent with the functional categories derived from the UniProt Knowledgebase database (Table 1).
Figure 3.
Altered proteins of B. pseudomallei cultured in LB broth at 42 °C under static conditions for 18 h. The proteins were categorised based on main biological functions by UniProt Knowledgebase (Swiss-Prot and TrEMBL entries) (A) and global protein network by STRING (B).
RNA expression of B. pseudomallei under heat stress conditions
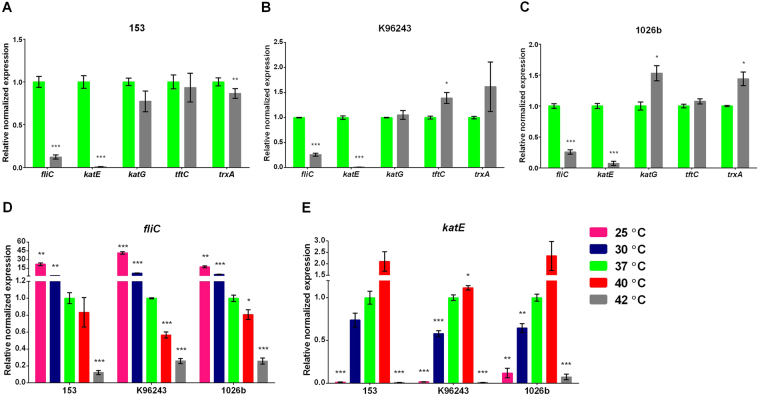
Of the multiple proteins altered by growth at 42 °C, redundancy in motility and antioxidant protein spots were observed among down-regulated proteins. To confirm these proteomic results, qRT-PCR was performed with three clinical isolates of B. pseudomallei from patients with melioidosis, strains 153, K96243 and 1026b for a motility gene (fliC), antioxidant protein-coding genes (katE, katG, tftC and trxA) and a reference gene (16S rDNA) (Fig. 4A–C). All three bacterial strains demonstrated significant down-regulation of fliC and katE expression at 42 °C compared with 37 °C, while reduction in katG, tftC and trxA expression was variable between the three strains. To further validate the effect of temperature, we analysed the expression of fliC, katE and 16S rDNA genes for B. pseudomallei cultured in LB broth at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C for 18 h. All three strains showed highest fliC expression at 25 °C followed by 30 °C, 37 °C, 40 °C and 42 °C (Fig. 4D). In contrast, these three isolates had highest katE expression at 37 °C and 40 °C and reduced katE expression at other temperatures (Fig. 4E).
Figure 4.
qRT-PCR relative expression levels of fliC, katE, katG, tftC and trxA in B. pseudomallei strains 153 (A), K96243 (B) and 1026b (C). (A), (B) and (C) B. pseudomallei was cultured in LB broth at 37 °C and 42 °C for 18 h. (D) and (E) B. pseudomallei was cultured in LB broth at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C for 18 h. 16S rDNA was used as a reference for the calculation of relative expression levels of other genes. The normalised expression levels were calculated by using 2−ΔΔCt method43. Data represent the mean, and error bars represent the standard deviation. *P < 0.05; **P < 0.01, ***P < 0.001, for 37 °C versus other temperatures.
Motility of B. pseudomallei at different temperatures
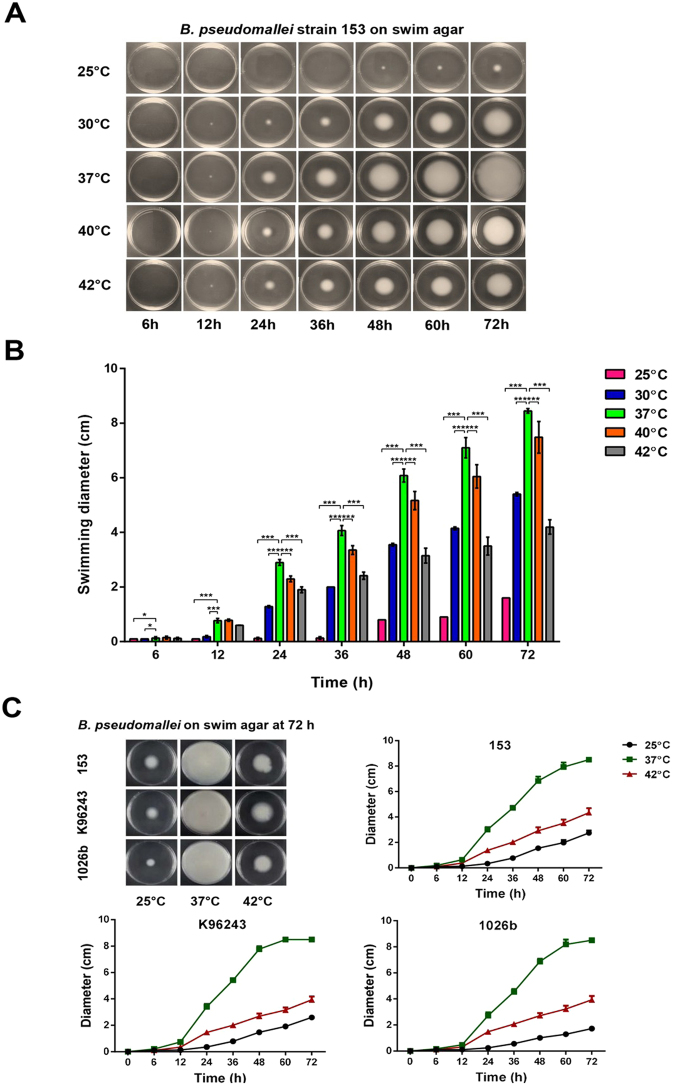
We hypothesised that B. pseudomallei reduces motility and anti-oxidative stress functions to maintain synthesis of vital proteins for growth at high temperatures. The thermal effect on swimming motility was investigated at a range of temperatures for strain 153. Swim plates were incubated at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C for 72 h. The zones of B. pseudomallei increased after incubation at all temperatures; the largest zone diameter was observed at 37 °C, followed by 40 °C, 42 °C and 30 °C, whereas the smallest zone diameter was observed at 25 °C (Fig. 5A,B). We re-examined swimming motility of bacteria at 25 °C, 37 °C and 42 °C for strains 153, K96243 and 1026b and obtained the same results (Fig. 5C). The swimming zone diameters of these strains increased overtime for all temperatures, with the largest diameters observed at 37 °C, followed by 42° and 25 °C. The diameters of all three strains after incubation at 25 °C and 42° were significantly lower than at 37 °C for all time points between 24 and 72 h (P < 0.001 for all strains, at 25 °C versus 37 °C and at 42 °C versus 37 °C).
Figure 5.
Swimming zone diameters of B. pseudomallei after incubation at different temperatures. (A) Swimming zones of B. pseudomallei strain 153 after incubation between 25 °C and 42 °C at different time points between 6 and 72 h, (B) Swimming zone diameters of B. pseudomallei strain 153 after incubation, (C) Swimming zone diameters of B. pseudomallei strains 153, K96243 and 1026b after incubation at 25 °C, 37 °C and 42 °C at different time points. The data represent the mean, and error bars represent the standard deviation.
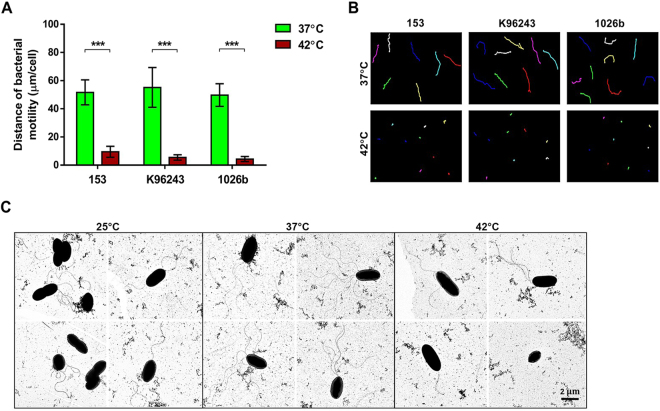
To determine whether the reduced swimming zone at 42 °C was a result of reduced motility or growth, video-assisted imaging under light microscopy was employed to examine motility of individual bacterial cells grown in LB broth for 18 h. The computational measurement data demonstrated that the average distance of bacterial motility of the three B. pseudomallei strains was significantly lower when incubated at 42 °C compared with 37 °C (Fig. 6A,B). In addition, B. pseudomallei cultured at 25 °C and 37 °C demonstrated forward directional movement, but when cultured at 42 °C showed rotating or circular movements. These data indicate that temperature affects the motility of B. pseudomallei.
Figure 6.
Live cell imaging analysis of individual B. pseudomallei cells for strains 153, K96243 and 1026b cultured at 37 °C and 42 °C for 6 h (A), and distance of bacteria cell motility (B). Motility was tracked for 20 s in 20 individual cells using ImageJ program (http://rsb.info.nih.gov/ij/). (C) Transmission electron microscopy of B. pseudomallei strain 153 cultured in LB broth at 25 °C, 37 °C and 42 °C for 6 h. ***P < 0.001.
Electron microscopy of B. pseudomallei under heat stress conditions
To understand the reduced swim motility of B. pseudomallei when cultured in LB broth for 6 h at 25 °C and 42 °C, we investigated the presence and number of flagella expressed by bacteria at these temperatures. Transmission electron microscopy confirmed a reduction in the proportion of flagellated bacteria at 42 °C (35%) compared with 37 °C (100%) and 25 °C (100%). The flagella of B. pseudomallei cultured at 42 °C appeared to be truncated. The mean (±standard deviation) number of flagella at 37 °C (3.3 ± 1.7, 95% CI 2.8–3.8) was significantly higher than those at 42 °C (1.6 ± 1.4, 95% CI 1.2–2.0) (P < 0.001) and 25 °C (2.1 ± 0.91, 95% CI 1.8–2.4) (P < 0.001) (Fig. 6C). These results showed that temperature changes affect the structure and expression of flagella in B. pseudomallei.
Susceptibility of B. pseudomallei to reactive oxygen intermediates at different growth temperatures
Proteome analysis and qRT-PCR of B. pseudomallei cultured at 42 °C demonstrated a pronounced decrease in expression of many antioxidant proteins. Because katE and katG encodes for catalases and peroxidase21–23, which are involved in H2O2 degradation, we tested the susceptibility of B. pseudomallei strain 153 to reactive oxygen intermediates (ROI) when cultured at different temperatures. The susceptibility of bacteria to ROI was investigated on LB agar plates containing 78 or 156 μM H2O2. Susceptibility varied depending on the growth temperature. B. pseudomallei cultured at 37 °C had the highest resistance to H2O2. In the presence of 78 μM H2O2, the most resistant bacteria were obtained from a culture at 37 °C followed by 40 °C, 25 °C and 30 °C. B. pseudomallei was most sensitive to H2O2 at 42 °C. The cell viability was low for bacteria cultured on the oxidant plate with 156 μM H2O2 (Fig. 7A). The same results were obtained for K96243 and 1026b. These data suggest that temperatures can regulate the expression of genes encoding antioxidant enzymes in B. pseudomallei, and that these enzymes may be involved in the resistance to ROI.
Figure 7.
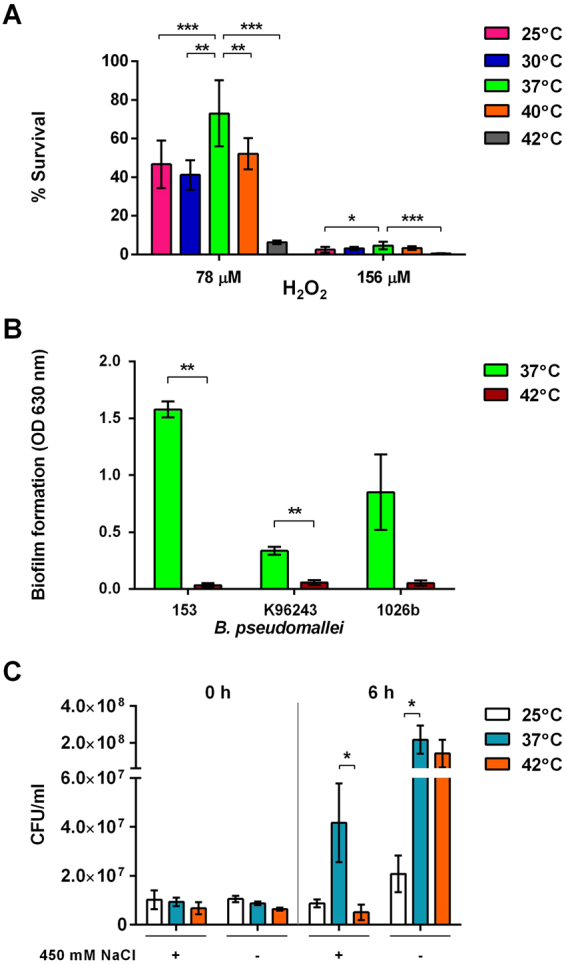
(A) Susceptibility of B. pseudomallei strain 153 to 78 µM and 156 µM H2O2 at different temperatures for 16 h. The number of colonies on oxidant agar plates containing H2O2 was normalised with those from plates without H2O2 and presented as the percentage of bacterial survival. (B) Biofilm formation of B. pseudomallei strains 153, K96243 and 1026b in LB broth for 48 h at 37 °C and 42 °C. The biofilm on a 96-well plate stained with 1% crystal violet was measured the OD at 630 nm. (C) Survival of B. pseudomallei under salt stress condition. B. pseudomallei were incubated at 25 °C, 37 °C and 42 °C for 18 h. The colony count was determined after exposure to 450 mM NaCl at 0 and 6 h. B. pseudomallei incubated at 25 °C, 37 °C and 42 °C in LB broth without added salt were used as the control. *P < 0.05; **P < 0.01, ***P < 0.001.
Biofilm-forming activity of B. pseudomallei
Biofilm formation has been previously reported to be associated with relapse in patients with melioidosis24. We, therefore, examined the effect of temperature on the biofilm-forming activity of B. pseudomallei strains 153, K96243 and 1026b in LB broth at 37 °C and 42 °C. Biofilm activity was significantly higher for all strains when cultured at 37 °C compared with 42 °C (Fig. 7B). The mean biofilm-forming activity varied between individual isolates. When cultured in LB broth at 42 °C, all strains failed to produce biofilm.
Susceptibility of B. pseudomallei to salt
We previously demonstrated that B. pseudomallei can increase thermal tolerance after exposure to a high concentration of NaCl13. In the present study, we further examined the susceptibility of bacteria to a high salt concentration following heat treatment for 6 h. Susceptibility of the three B. pseudomallei strains to salt stress was tested with pre-incubation at 25 °C, 37 °C and 42 °C for 18 h prior to exposure to 450 mM NaCl in LB broth at the same temperatures for 6 h. The average numbers of viable bacteria at all temperatures were lower for bacteria cultured in LB broth with high salt compared with the bacteria cultured in LB without added salt as the control (Fig. 7C). In LB broth with high NaCl concentration, the average number of viable bacteria was highest for bacteria cultured at 37 °C with a bacterial count of 4.17 × 107 CFU/ml, which was significantly different from bacteria cultured at 42 °C with a bacterial count of 5.06 × 106 CFU/ml (P = 0.047). In LB broth without added salt as the control, the average number of viable bacteria cultured at 37 °C was not significantly different from that of 42 °C (2.17 × 108 CFU/ml versus 1.42 × 108 CFU/ml, P = 0.102) but was significantly higher than that of 25 °C (2.08 × 107 CFU/ml, P = 0.049). These data confirm the effect of temperature on the resistance of B. pseudomallei to salt stress.
Discussion
Melioidosis is a public health burden in tropical countries and an emerging disease that is estimated to cause 89,000 deaths globally each year2. Patients with melioidosis present with abscess and protracted fever that requires prolonged treatment. Our study focused on the effect of temperatures that are encountered by B. pseudomallei in the environment and the human host during infection. Our data demonstrated that B. pseudomallei could grow and survive in a wide range of temperatures. Thermal changes were found to trigger several responses in B. pseudomallei at the proteomic level. The proteins produced by B. pseudomallei due to thermic change included metabolic enzymes, chaperones, cell motility proteins, antioxidants, and structural proteins. These proteins were particularly produced by B. pseudomallei at 37 °C, a temperature which was found to support bacterial growth, motility, biofilm formation and resistance to oxidative stress.
The rapid growth of B. pseudomallei at 37 °C suggests that B. pseudomallei may proficiently replicate upon entry into hosts. The ability of the bacteria to grow at 38 °C–40 °C suggests that B. pseudomallei are capable of replicating in a host with a high fever. The range of temperatures used here of 25 °C–42 °C are similar to environmental conditions in tropical regions that support B. pseudomallei in soil and water. Indeed, it has been found that B. pseudomallei can remain viable with high bacterial numbers of more than 10,000 CFU per gram of soil in northeast Thailand3,4,25. Although the growth of B. pseudomallei is reduced at lower temperatures such as 25 °C, stationary phase is finally achieved and cell viability remains high. However, our study demonstrated that B. pseudomallei grew at a high rate at 42 °C but viability slowly declined after stationary phase if the high temperature was maintained. Our results suggested that the dynamics of temperature changes during the day and night or in different seasons may affect the growth of B. pseudomallei, but that the bacterium can tolerate fluctuation in environmental temperature in tropical climates.
Different growth temperatures had a broad impact on protein expression by B. pseudomallei. Proteomic analysis identified up- and down-regulated proteins in response to temperature changes, suggesting that temperature is an important environmental signal for cellular metabolism aimed at maintaining the growth and biological activity of the bacterium. Many studies have shown that alteration of bacterial cellular proteins is a necessary mechanism for cell survival under heat stress15,16,26. During early exposure to high temperatures, reactive oxygen species (ROS) are generated, and there is an accumulation of misfolded membrane proteins. Our result demonstrated that at 42 °C, B. pseudomallei had reduced levels of outer membrane proteins (OmpA) and chitin-binding proteins (CBPs). OmpA is a structural and immunogenic protein27,28 and CBPs of many bacterial pathogens contain surface-exposed domains that support infection of non-chitinous mammalian hosts29. The increased levels of many chaperones at high temperatures are consistently observed in other bacteria17,19. Heat shock proteins (HSPs) are well known to participate in protein folding, refolding and removal of non-functional damaged proteins16,18. Escherichia coli respond to high temperatures by activating a heat shock response. The stability of mRNAs of heat shock genes has been shown to be maintained by the cold shock protein C (CspC)15, but this mechanism is still unknown for B. pseudomallei.
An adaptive mechanism utilised by bacteria to survive at elevated temperatures is the reduction of metabolic activity, which is crucial for energy conservation30. Our findings confirmed that expression of several metabolic proteins including acetoacetyl-CoA reductase, D-beta-hydroxybutyrate dehydrogenase, ATP synthase, elongation factor Tu, non-ribosomally encoded peptide/polyketide synthase, phosphoglycerate kinase, inorganic pyrophosphatase, rhodanese-related sulfotransferase and carbamate kinase were decreased. We previously showed that carbamate kinase encoded by arcC is a component of the arginine deiminase system, which facilitates acid tolerance in B. pseudomallei12. The decreased level of carbamate kinase under heat stress would potentially lead to defective cell survival processes. However, some cellular metabolic proteins were upregulated, including carboxypeptidase protein, pyruvate aminotransferase and phosphoserine aminotransferase.
In our study, qRT-PCR analysis confirmed the proteomic results in the modification of transcription levels of fliC (encoding flagellin) and katE expression by temperature changes. Experiments on three B. pseudomallei strains using swim agar validated the proteomic results that motility and the presence of flagella were affected by temperature change, with the highest function observed at 37 °C. Temperature-dependent regulation of flagella expression was previously reported in B. thailandensis, wherein bacterial growth at 28 °C enhanced cell motility and flagella expression through a mechanism involving the regulation of the fliC gene at the mRNA stability level31. B. thailandensis is a non-virulent environmental species of Burkholderia which is closely related to B. pseudomallei. We observed similar transcriptomic results in B. pseudomallei that flicC expression was highly expressed at 25 °C. However, our results were based on three B. pseudomallei strains that showed larger swimming zones at 37 °C compared with those at 25 °C and 30 °C. The discrepancy between transcriptomic results and swimming zone at 25 °C and 30 °C may be associated with bacterial growth rate or a complex process of a translation inhibition of FliC, an issue that requires further investigation. Our data from live cell imaging and electron microscopy also confirmed that the large zone diameter at 37 °C was associated with individual cell movement and flagella production. However, swimming motility was reduced when B. pseudomallei were cultured at 42 °C, which was consistent with proteomic and mRNA transcription of the fliC gene. The proteomic analysis suggested that response to increased temperature may enable bacterial cells to produce more important stress response proteins, namely chaperones and metabolic enzymes and antioxidants. Temperature has been reported to regulate flagellar motility genes in other bacteria such as Pseudomonas syringae32 and Listeria monocytogenes33.
We also found that B. pseudomallei underwent a number of phenotypic adaptations under different growth temperatures. Altered flagella expression could be linked to the virulence and modulation of the immune response during infection. French et al. demonstrated that bacterial flagella support rapid intracellular motility and efficient cell-to-cell spread, thus forming multinucleate giant cells (MNGC), leading to cell death34. A notable virulence feature of B. pseudomallei is the formation of MNGC, which occurs in many cell types including phagocytic and non-phagocytic cells35,36. Flagellin (FliC) of B. pseudomallei is an important innate immune ligand for Toll-like receptor (TLR) 5. It activates NF-κB and leads to the production of inflammatory cytokines37. Our data highlight the importance of flagellin expression, which may be transiently regulated by the alteration of host temperature.
All three B. pseudomallei strains were efficiently resistant to ROI, with the highest resistance observed when cultured at 37 °C. This observation on oxidant agar was consistent with the results of proteomic and transcriptional analyses showing high expression of katE at 37 °C and 42 °C compared with those at 42 °C and other temperatures. katE (BPSS2214) and katG (BPSL2865) encode for catalase and peroxidase enzymes, respectively, which catalyse H2O2 degradation. These enzymes are important for the survival of facultative aerobic organisms exposed to oxidative stress. katG and katE expression in B. pseudomallei has also been reported to be regulated by rpoS and oxyR38, whereas katE expression is regulated by rpoN239 in response to several stress conditions, such as nutritional starvation and oxidative stress. Further studies are required to investigate the effect of temperature on the expression and roles of these regulators.
Our data demonstrated that biofilm activity was significantly higher for all three strains when cultured at 37 °C compared with 42 °C. Consistent with our study, the effect of temperature on biofilm formation has recently been reported in B. pseudomallei, in which the level of biofilm formation by B. pseudomallei strain 1026b in LB medium decreased ∼6-fold at 40 °C and 42 °C compared with 37 °C40. The role of temperature in the regulation of biofilm formation has been linked to cyclic di-GMP signaling in B. pseudomallei40. Our data suggest that biofilm formation is associated with B. pseudomallei cells that are more metabolically active during growth at 37 °C. The decrease in biofilm seen at 42 °C was possibly related to the down-regulation of the flagella expression and other metabolic enzymes.
We previously showed that a high NaCl concentration affects the survival and adaptive mechanisms of B. pseudomallei. In response to exposure to high NaCl concentration, B. pseudomallei isolates show an increased thermal tolerance, oxidative resistance, and plaque-forming efficiency13. However, the result of the present study demonstrated that B. pseudomallei had the greatest tolerance to salt stress following pre-incubation at 37 °C in comparison with that at 25 °C and 42 °C. Increased temperature of 42 °C was negatively associated with reduced bacterial viability under the same salt stress conditions. Resistance to salt at 37 °C may provide a major benefit for B. pseudomallei stability in environmental niches where the bacteria encounter a warm temperature but high salt concentrations.
Our study suggested that the human body temperature of 37 °C facilitates maximum growth rate, motility and anti-oxidative stress functions in B. pseudomallei. These mechanisms may be a prerequisite for acute infection and dissemination. In conditions with high temperatures, B. pseudomallei expresses a number of stress-response proteins but decreases its motility by reducing flagella expression. This may ultimately lead to latent infection if the patients do not recover from melioidosis.
Materials and Methods
Ethics statement
The study was approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (approval number: MUTM2018-009-01). All bacterial isolates obtained from humans were anonymous.
B. pseudomallei strains
Three B. pseudomallei isolates from patients with melioidosis in northeast Thailand were used: strains 153, 1026b and K96243. All isolates grew with a type I colony morphology on Ashdown agar after incubation for 4 days at 37 °C in air41. Unless otherwise stated, a single colony of B. pseudomallei was cultured on LB, Lennox agar or broth and incubated at 37 °C in air for 18 h before being used in each experiment.
Bacterial growth curve analysis
Bacterial culture was performed in a BSL-3 laboratory. Several colonies of B. pseudomallei were suspended in sterile PBS and the optical density (OD) at 600 nm adjusted to obtain a bacterial concentration of approximately 1 × 108 colony-forming units (CFU)/ml. Five microliters of bacterial suspension was added to 5 ml of LB broth to make a final concentration of 1 × 105 CFU/ml. Cultures were then incubated in duplicate at five different temperatures under static conditions: 25 °C, 30 °C, 37 °C, 40 °C and 42 °C and sampled at time intervals (0-, 6-, 12-, 24-, 36-, 48-, 60- and 72 h), aliquots from which were serially diluted in sterile PBS. Ten microliters of each dilution were inoculated on Columbia agar in triplicate, followed by incubation at 37 °C in air for 16 h before a colony count was performed.
Protein extraction
A single colony of B. pseudomallei on Ashdown agar was picked and inoculated into 100 ml of LB broth, which was incubated at 37 °C or 42 °C under static conditions for 18 h. Two independent experiments were performed. Bacteria were harvested by centrifugation at 4,500 × g for 30 min followed by washing with PBS. The bacterial pellet was resuspended in 1 ml of a cold lysis buffer (5 mM EDTA and 1 mM phenylmethylsulfonyl fluoride)12 and then sonicated on ice at 22% amplitude for 3 min. The cell lysate was centrifuged at 14,000 × g at 4 °C for 3 min, and the supernatant was then collected and filtered using a 0.2-μm filter. Protein samples were stored at −80 °C until use.
2D gel electrophoresis and quantitative spot intensity analysis
After cleaning the protein samples, protein concentration was measured using a 2D quantification kit (GE Healthcare Bio-Sciences). Protein complexes (700 µg) derived from two independent experiments for each condition were separated by isoelectric focusing on Immobiline DryStrip (linear pH gradient of 4–7, 18 cm) followed by 12% polyacrylamide gel electrophoresis, as previously described12. Separated protein spots were visualised with colloidal Coomassie blue G-250 stain and then captured using an Image Scanner II and LabScan software version 5.0 (GE Healthcare Bio-Sciences). Image Master 2D Platinum software 7.0 (GE Healthcare Bio-Sciences) was used for matching and analysis of the protein spots in 2D gels. The fold change in the intensity of matched protein spots between 37 °C and 42 °C was calculated. The differential spots representing more than a two-fold change were then subjected to protein identification by mass spectrometry.
In-gel tryptic digestion and protein identification by MALDI-TOF MS
Candidate proteins were excised from the gel using the Ettan Spot Handling Workstation (GE Healthcare Bio-Sciences) followed by digestion with trypsin, as previously described12. The trypsinized samples were deposited onto the 96-well MALDI target plate (MTP 384 polished steel TF), and protein mass spectra were obtained using an Autoflex MALDI-TOF MS (Bruker Daltonik, Bremen, Germany).
MASCOT searching and protein interaction network analysis
The MS data were exported as searchable files and used to search against peptide mass fingerprint in the MASCOT database (http://www.matrixscience.com). Protein identification was achieved using a protein BLAST search. All identified proteins were classified based on their main biological functions using UniProt Knowledgebase (Swiss-Prot and TrEMBL entries). The differentially expressed proteins isolated at 42 °C compared with at 37 °C were further analysed for protein interaction network using a STRING tool software (version 8.3) (http://string.embl.de/)42. Predicted protein–protein associations were queried through experimentally derived physical protein interactions from the research literature and databases of curated biological pathway knowledge.
RNA extraction
B. pseudomallei strains K96243, 153 and 1026b were inoculated into 5 ml LB broth and incubated at 25 °C, 30 °C, 37 °C, 40 °C and 42 °C for 18 h under static conditions, after which bacteria were harvested by centrifugation at 12,000 rpm for 2 min. The pellet was washed with 1 ml of PBS, and RNA was extracted using RNeasy kit (Qiagen, Germany). The integrity of the purified RNA was examined by agarose gel electrophoresis, and the concentration was measured using a Nanodrop spectrophotometer (Thermo Scientific, USA). Contaminant genomic DNA was removed using DNase I treatment according to the manufacturer’s instructions (Thermo Scientific, USA), and the presence of residual DNA was checked by PCR using primers for the 16S rDNA gene (Table S1).
Quantitative reverse transcriptase PCR (qRT-PCR)
Two-step qRT-PCR was used to quantitatively measure gene expression using the iScriptTM Reverse Transcription Supermix for RT-qPCR (Bio-Rad, USA) and a CFX96 Touch TM Real-Time PCR Detection System with Bio-Rad CFX Manager software version 3.0 (Bio-Rad). All primer pairs were designed using NCBI PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer sequences are shown in Table S1. The amplification was performed for the fliC, katE, katG, tftC, trxA and 16 S rDNA genes in duplicate in 10 µl total volume containing 5 µl of 2 iTaq Universal SYBR Green (Bio-Rad), 1 µl of cDNA, 0.4 µl of each primer (10 µmol/l), and 3.2 µl of distilled water. The cycle conditions were as follows: 1 cycle of 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. After amplification, melting curve analysis was conducted by increasing the annealing temperature by 0.1 °C per step from 65 °C to 95 °C. The 16 S gene was used as the reference for calculating the relative expression levels of other genes. The normalised expression levels were calculated by using the 2−ΔΔCt method, where ΔΔCt = (Ct target gene − Ct 16S rDNA gene) at other temperatures − (Ct target gene − Ct 16S rDNA gene) at 37 °C43.
Swimming motility assay
Two single colonies of B. pseudomallei were cultured in 5 ml of LB broth at a range of temperatures (25 °C, 30 °C, 37 °C, 40 °C and 42 °C) under static conditions for 18 h. The OD at 600 nm of the bacterial suspension was adjusted in sterile PBS to a concentration of 1 × 108 CFU/ml and point inoculated into the centre of swimming agar plate (1% tryptone, 0.5% NaCl and 0.3% agar) using a sterile toothpick, as described previously41. The swim plates were further incubated at the same temperatures. The swimming zone diameters were measured in three different positions including the widest, medium and shortest diameters of the circular turbid zone formed by the bacterial cells migrating away from the point of inoculation at 0-, 6-, 12-, 24-, 36-, 48-, 60- and 72 h time intervals and the average zone diameters are presented.
Live cell imaging of cell motility in B. pseudomallei
Live cell imaging was used to monitor the motility of B. pseudomallei using the hanging drop method. B. pseudomallei were cultured on Ashdown agar at 37 °C overnight. Colonies of each isolate were harvested and suspended in 10 ml of LB broth, and their OD at 600 nm was adjusted to a concentration of 1 × 1010 CFU/ml. Equal volumes of the bacterial suspension (2.5 ml) were added into two tubes containing LB broth to make a final volume of 5 ml and incubated separately at 37 °C and 42 °C for 6 h under static conditions to obtain log-phase cultures. Thereafter, 30 µl of the bacterial suspension was dropped on a glass slide with a depression, over which a cover slip was placed. Motility was observed using a light microscope (Leica DM750, Wetzlar, Germany) at 1000 × magnification. A video of the motility was recorded and the motility of 20 individual cells tracked for 20 s using ImageJ program (http://rsb.info.nih.gov/ij/) with the manual tracking plug-in.
Transmission electron microscopy
B. pseudomallei strain 153 was cultured in LB broth at 25 °C, 37 °C and 42 °C for 6 h. A preparation of the bacterial cells was examined using transmission electron microscopy (TEM) for the presence of flagella, as previously described44. The bacterial cells were observed with a Hitachi Electron Microscope HT-7700 (Hitachi, Japan). The presence of bacterial flagella was recorded for 50 bacteria in each temperature.
Determination of susceptibility of B. pseudomallei to reactive oxygen intermediates (ROI)
The susceptibility of B. pseudomallei to ROI at different temperatures was determined on oxidant agar plates, as previously described7. Briefly, B. pseudomallei were inoculated in 10 ml of LB broth and incubated at different temperatures (25 °C, 30 °C, 37 °C, 40 °C and 42 °C) for 16 h. The bacterial suspension from each condition was diluted with sterile PBS to make 10-fold dilutions. Ten microliters of a serially diluted inoculum were inoculated onto LB agar containing 0, 78 and 156 μM H2O2 in triplicate. The colonies were counted after incubation at 37 °C in air for 24 h. The number of colonies on the plates containing H2O2 was normalised with those from plates without H2O2 by presenting as the percentage of bacterial survival.
Assessment of biofilm-forming activity of B. pseudomallei
The biofilm-forming activity of B. pseudomallei was assessed as described previously41. Briefly, the bacteria were inoculated into 10 ml of LB broth and incubated at 37 °C with shaking for 18 h. The OD at 600 nm of the bacterial suspension was adjusted to 1.0 with fresh LB broth and 200 µl of the suspension was added into a 96-well flat-bottomed plastic tissue culture plate (FALCON, USA), with eight wells/strain. Wells with only LB broth were included as negative controls. The plates were incubated at 37 °C or 42 °C to allow adhesion for 3 h. The supernatant was gently aspirated and replaced with 200 µl of fresh LB broth, and the plate was continually incubated at the same temperature for an additional 21 h. The supernatant was replaced with a fresh LB broth and incubated as above for an additional 24 h. Thereafter, the wells were washed with PBS, fixed with methanol for 15 min, and air-dried at room temperature. The plates were stained with 1% crystal violet and solubilised with 250 µl of 33% (v/v) glacial acetic acid per well before measuring the OD at 630 nm using a TECAN microplate reader (TECAN, Switzerland).
Salt stress assay
The survival of B. pseudomallei strains K96243, 1026b and 153 under salt stress conditions were determined by observing the number of viable bacteria after exposure to 450 mM NaCl, as previously described13. Briefly, B. pseudomallei were incubated in 5 ml of LB broth at 25 °C, 37 °C and 42 °C for 18 h. The bacterial cells were harvested, washed and resuspended in PBS. The OD at 600 nm of the bacterial suspension was adjusted to 0.15 to obtain a bacterial concentration of 1 × 108 CFU/ml. The bacteria were then inoculated at a dilution of 1:10 into 10 ml of LB broth containing 450 mM NaCl or LB broth without added salt as the control. The bacteria were further incubated at the same temperatures for 6 h with shaking. A 10-fold dilution of the bacteria was prepared and plated onto LB agar. After incubation at 37 °C for 24 h, colonies were counted. The mean numbers of colonies of the three B. pseudomallei strains were presented as the CFU.
Statistical analysis
Statistical analyses were performed using Stata, version 12 (StataCorp LP, College Station, TX, USA). One-way ANOVA or Student t-test was used to test differences in quantitative data among different groups. All data in this study were presented as mean ± standard deviation. Differences were considered statistically significant at a P value < 0.05.
Data availability
All data generated or analysed during this study are included in this published article.
Electronic supplementary material
Acknowledgements
We are grateful for the support from staff at the Department of Microbiology and Immunology and at the Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine Mahidol University. We would like to thank Aunchalee Thanwisai, Amporn Rungruengkitkun and Sarunporn Tandhavanant for their technical assistance. This study was funded to N.C. by a Wellcome Trust Career Development award in Public Health and Tropical Medicine, UK (grant 087769/Z/08/Z) (http://www.wellcome.ac.uk) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number U01AI115520 (https://www.nih.gov). S.P. and N.C. were supported by an ICTM grant from the Faculty of Tropical Medicine, Mahidol University. P.P. was supported by a research grant from the Faculty of Tropical Medicine, Mahidol University, Fiscal Year 2014. The funder had no role in study design and interpretation, the decision to publish or preparation of the manuscript.
Author Contributions
S.P., K.S., T.Y., P.P. and N.C. performed experiments and analysed the data. N.C. designed the experiment and supervised the study. N.C. and S.J.P. formulated the hypothesis and wrote the manuscript. All authors reviewed and approved the final draft.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27356-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Eng J Med. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 3.Chantratita N, et al. Genetic diversity and microevolution of Burkholderia pseudomallei in the environment. PLoS Negl Trop Dis. 2008;2:e182. doi: 10.1371/journal.pntd.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuthiekanun V, et al. Burkholderia pseudomallei is genetically diverse in agricultural land in Northeast Thailand. PLoS Negl Trop Dis. 2009;3:e496. doi: 10.1371/journal.pntd.0000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis. Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pumpuang A, et al. Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans R Soc Trop Med Hyg. 2011;105:598–600. doi: 10.1016/j.trstmh.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tandhavanant S, et al. Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiol. 2010;10:303. doi: 10.1186/1471-2180-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone JK, et al. Structural characterization of polysaccharides expressed by Burkholderia oklahomensis E0147. Carbohydr Res. 2014;386:68–72. doi: 10.1016/j.carres.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Valade E, et al. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J Bacteriol. 2004;186:2288–2294. doi: 10.1128/JB.186.8.2288-2294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton RE, et al. Quorum sensing negatively regulates multinucleate cell formation during intracellular growth of Burkholderia pseudomallei in macrophage-like cells. PloS One. 2013;8:e63394. doi: 10.1371/journal.pone.0063394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burtnick MN, et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. 2011;79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chantratita N, et al. Proteomic analysis of colony morphology variants of Burkholderia pseudomallei defines a role for the arginine deiminase system in bacterial survival. J Proteomics. 2012;75:1031–1042. doi: 10.1016/j.jprot.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pumirat, P. et al. Effects of sodium chloride on heat resistance, oxidative susceptibility, motility, biofilm and plaque formation of Burkholderia pseudomallei. Microbiologyopen6, 10.1002/mbo3.493 (2017). [DOI] [PMC free article] [PubMed]
- 14.Burtnick MN, Brett PJ. Burkholderia mallei and Burkholderia pseudomallei cluster 1 type VI secretion system gene expression is negatively regulated by iron and zinc. PloS One. 2013;8:e76767. doi: 10.1371/journal.pone.0076767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenhar Y, Rasouly A, Biran D, Ron EZ. Adaptation of Escherichia coli to elevated temperatures involves a change in stability of heat shock gene transcripts. Environ Microbiol. 2009;11:2989–2997. doi: 10.1111/j.1462-2920.2009.01993.x. [DOI] [PubMed] [Google Scholar]
- 16.Murata M, et al. Molecular strategy for survival at a critical high temperature in Eschierichia coli. PloS One. 2011;6:e20063. doi: 10.1371/journal.pone.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 18.Ryabova NA, Marchenkov VV, Marchenkova SY, Kotova NV, Semisotnov GV. Molecular chaperone GroEL/ES: unfolding and refolding processes. Biochemistry (Mosc) 2013;78:1405–1414. doi: 10.1134/S0006297913130038. [DOI] [PubMed] [Google Scholar]
- 19.Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratkowsky DA, Olley J, Ross T. Unifying temperature effects on the growth rate of bacteria and the stability of globular proteins. J Theor Biol. 2005;233:351–362. doi: 10.1016/j.jtbi.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Heym B, Zhang Y, Poulet S, Young D, Cole ST. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol. 1993;175:4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewen PC. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng M, et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limmathurotsakul D, et al. Role of Burkholderia pseudomallei biofilm formation and lipopolysaccharide in relapse of melioidosis. Clin Microbiol Infect. 2014;20:O854–856. doi: 10.1111/1469-0691.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limmathurotsakul D, et al. Burkholderia pseudomallei is spatially distributed in soil in northeast Thailand. PLoS Negl Trop Dis. 2010;4:e694. doi: 10.1371/journal.pntd.0000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noor R. Mechanism to control the cell lysis and the cell survival strategy in stationary phase under heat stress. Springerplus. 2015;4:599. doi: 10.1186/s40064-015-1415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nithichanon A, et al. Boosting of post-exposure human T-cell and B-cell recall responses in vivo by Burkholderia pseudomallei-related proteins. Immunology. 2017;151:98–109. doi: 10.1111/imm.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara Y, Mohamed R, Nathan S. Immunogenic Burkholderia pseudomallei outer membrane proteins as potential candidate vaccine targets. PloS One. 2009;4:e6496. doi: 10.1371/journal.pone.0006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frederiksen RF, et al. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology. 2013;159:833–847. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez J, Premier GC, Guwy AJ, Dinsdale R, Kleerebezem R. Metabolic models to investigate energy limited anaerobic ecosystems. Water Sci Technol. 2009;60:1669–1675. doi: 10.2166/wst.2009.224. [DOI] [PubMed] [Google Scholar]
- 31.Peano C, et al. Gene and protein expression in response to different growth temperatures and oxygen availability in Burkholderia thailandensis. PloS One. 2014;9:e93009. doi: 10.1371/journal.pone.0093009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hockett KL, Burch AY, Lindow SE. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PloS One. 2013;8:e59850. doi: 10.1371/journal.pone.0059850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamp HD, Higgins DE. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 2011;7:e1002153. doi: 10.1371/journal.ppat.1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French CT, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc Natl Acad Sci USA. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteley, L. et al. Entry, intracellular survival, and multinucleated-giant-cell-forming activity of Burkholderia pseudomallei in human primary phagocytic and nonphagocytic cells. Infect Immun85, 10.1128/IAI.00468-17 (2017). [DOI] [PMC free article] [PubMed]
- 36.Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun. 2000;68:5377–5384. doi: 10.1128/IAI.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chantratita N, et al. Screen of whole blood responses to flagellin identifies TLR5 variation associated with outcome in melioidosis. Genes Immun. 2014;15:63–71. doi: 10.1038/gene.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jangiam W, Loprasert S, Smith DR, Tungpradabkul S. Burkholderia pseudomallei RpoS regulates OxyR and the katG-dpsA operon under conditions of oxidative stress. Microbiol Immunol. 2010;54:389–397. doi: 10.1111/j.1348-0421.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 39.Diep DT, Phuong NT, Hlaing MM, Srimanote P, Tungpradabkul S. Role of Burkholderia pseudomallei sigma N2 in amino acids utilization and in regulation of catalase E expression at the transcriptional level. Int J Bacteriol. 2015;2015:623967. doi: 10.1155/2015/623967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plumley, B. A. et al. Thermoregulation of biofilm formation in Burkholderia pseudomallei is disrupted by mutation of a putative diguanylate cyclase. J Bacteriol199, 10.1128/JB.00780-16 (2017). [DOI] [PMC free article] [PubMed]
- 41.Chantratita N, et al. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol. 2007;189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Ngamdee W, et al. Competition between Burkholderia pseudomallei and B. thailandensis. BMC Microbiol. 2015;15:56. doi: 10.1186/s12866-015-0395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.