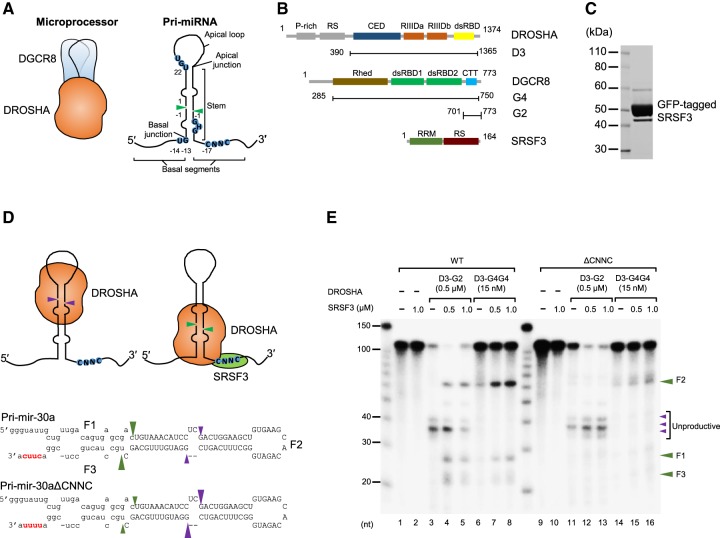
FIGURE 1.
SRSF3 recruits DROSHA to the CNNC-containing basal junction. (A) Microprocessor is a trimeric complex consisting of an RNase III DROSHA and a DGCR8 dimer. The typical pri-miRNA, a substrate of the Microprocessor complex, has a stem–loop structure. The green arrowheads indicate the DROSHA cleavage sites. The basal junction and the basal UG motif located at 14 nt from the cleavage site are recognized by DROSHA. The apical UGU motif of the apical loop that appeared at 22 nt from the cleavage site is recognized by DGCR8. The CNNC motif of the basal segment that resides at 17 nt from the cleavage site interacts with SRSF3. (B) Protein constructs used in this study. The first and end residue numbers for each construct are shown. (P-rich) Proline-rich domain, (RS) arginine/serine-rich domain, (CED) central domain, (RIIIDa and RIIIDb) RNase III domains, (dsRBD) dsRNA-binding domain, (Rhed) RNA-binding heme domain, (CTT) C-terminal tail region, (RRM) RNA recognition motif. SRSF3 is composed of 164 amino acids and its molecular weight is about 19 kDa. It has one RRM at the N terminus and one RS domain at the C terminus with extensive phosphorylation on serine residues. (C) The purity of the GFP-tagged SRSF3 on SDS–PAGE. (D) The pri-mir-30a substrates. The uppercase letters represent the pre-miRNA region. The CNNC and ΔCNNC motifs are highlighted in red. The green and purple arrowheads indicate the productive and unproductive cleavage sites of DROSHA, respectively. (E) Processing of the pri-mir-30a substrates by D3–G2 or D3–G4G4 and SRSF3. The internally labeled pri-mir-30a substrates were incubated with the indicated proteins for 1 h under the conditions described in Materials and Methods. The three products, a 5′-fragment, pre-miRNA, and a 3′-fragment were named as F1, F2, and F3, respectively.