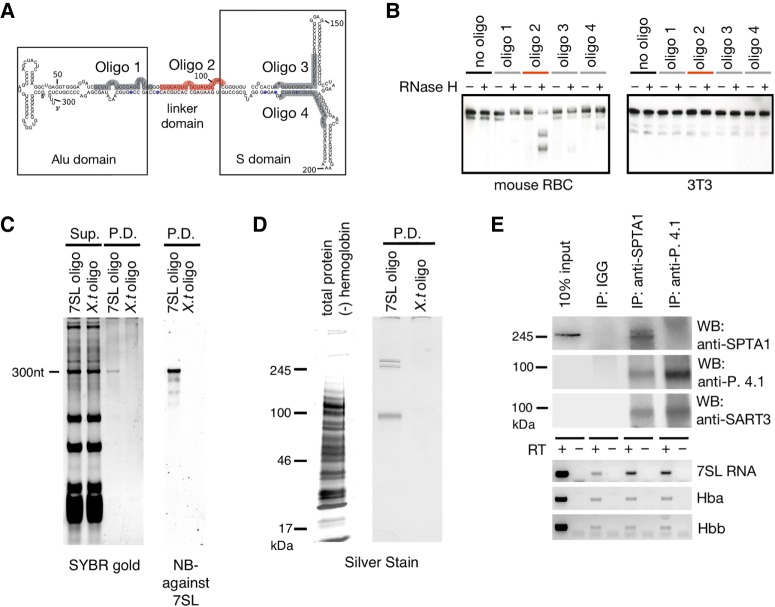
FIGURE 4.
Affinity purification of 7SL RNA. (A) Sequence of the four antisense oligonucleotides used to probe 7SL RNA in RBCs. (B) 7SL RNA from an RBC lysate was cleaved by RNase H only when it was hybridized to oligo 2 (red). 7SL RNA from mouse 3T3 cells was not cleaved. Experiment was done twice. (C) Affinity purification of 7SL. Total RNA in the supernatant (Sup.) and the pull-down (P.D.) after hybridization with a 7SL RNA antisense probe (7SL oligo) or a control probe (X.t. oligo). RNA was detected by SYBR gold. Analysis of the pulled-down RNA showed readily detectable 7SL RNA by Northern blotting. Experiment was done twice. (D, left lane) Total proteins (minus hemoglobins) before pull-down. (Right lanes) Proteins in the pull-down fractions shown in C. Proteins detected by silver staining. In the 7SL pull-down fraction, the major bands are assumed to be spectrin (α and β chains), protein 4.1 and band3, based on their molecular weights and on the abundance of these proteins in the mass spectroscopic analysis. Experiment was done twice. (E) Proteins and RNA pulled down by antibodies against spectrin α and protein 4.1. (Top panels) Pulled-down proteins were analyzed by Western blots against spectrin α, protein 4.1 and SART3. SART3 was detected in both pull-downs. Only spectrin alpha was sufficiently abundant to be detected in the input lane. Western blots were performed once. (Bottom panels) Pulled-down RNAs were analyzed by RT-PCR using oligos targeting 7SL RNA and two hemoglobin mRNAs, Hba and Hbb. 7SL RNA was detected in both pull-downs. Northern blots were done twice.